An Everyday DNA blog article
Written by: Sarah Sharman, PhD, Science writer
Illustrated by: Cathleen Shaw
As summer comes to an end and we grow nearer to the spooky season, I have already begun planning out my scary movie schedule. I love the scenes where a group of campers are sitting around a campfire in the middle of a dark wooded area late at night regaling ghost stories. Usually right at the climax of the story, a startling noise comes from the woods making the campers believe that there is actually a ghost in their presence.
In folklore, a ghost is the soul or spirit of a deceased person or animal that appears to the living. Ghosts have been portrayed in pop culture as children wearing sheets with holes cut for their eyes, transparent humanlike specters, cartoon ghosts like Casper and the Pac-Man ghosts, big green phantoms like Slimer, and zombie-esque ghosts like Beetlejuice.
Ghosts do not just show up to haunt us in pop culture, it turns out they also show up in our ancestry. You are probably thinking, wait a second there are ghosts in my family tree? I don’t mean that the ghosts of your ancestors are going to show up for Thanksgiving dinner. But by using DNA sequencing technology, scientists have discovered several “ghost” species that do not exist in the fossil records. Let’s learn more about this phenomenon called a ghost lineage.
What is a ghost lineage?
Evolution by its simplest definition is change over time. It is responsible for shaping the tree of life and providing the incredible array of diversity scientists see both in fossil records and in modern ecosystems. As organisms evolve and genetic modifications are inherited, their evolutionary paths diverge from their ancestors. This produces a branching pattern of evolutionary relationships.
Phylogenetics is the study of these evolutionary relationships. Diagrams that represent scientists’ best hypothesis about how a set of species evolved from a common ancestor are called phylogenetic trees. When humans map out their family tree, they usually have written documents or photographs to help them identify relatives. Evolutionary lineages of other species are not always as easy to uncover.
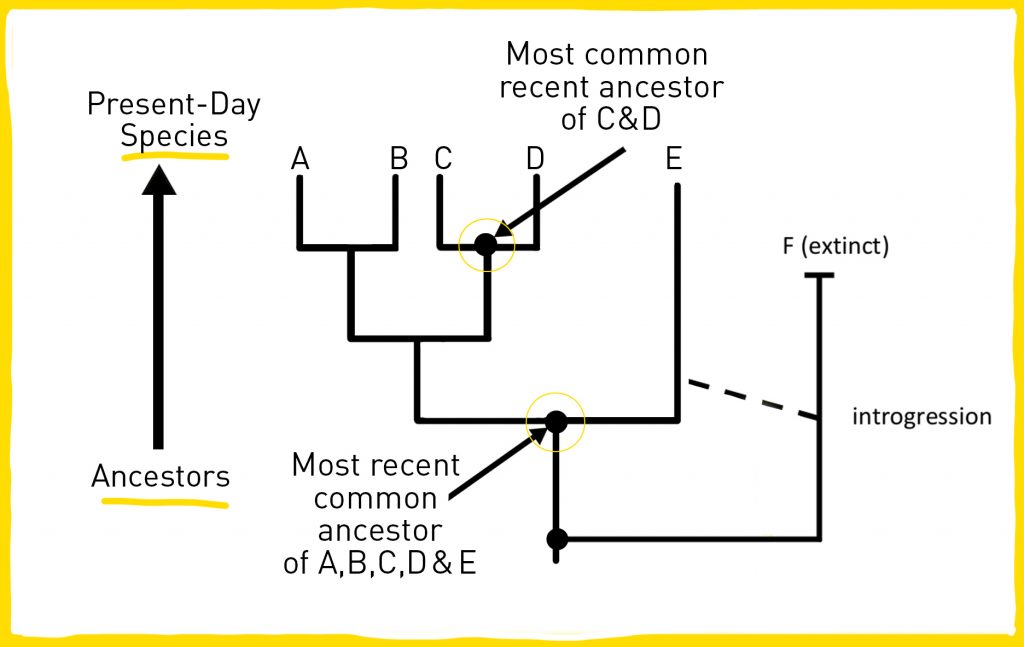
To build a phylogenetic tree, scientists collect data about the characteristics of each organism they are interested in. These characteristics could include physical characteristics (also called morphologic characteristics), genetic sequences, and behavioral traits. The species or groups of interest are shown at the tips of the tree’s branches (represented as A, B, C, D, and E in the example below).
Scientists organize species into groups based on shared characteristics (like having four-limbs, wings, or shared DNA sequences). By grouping the organisms into less and less inclusive categories, scientists build a tree having the earliest common ancestor at the trunk, or base, and each species of interest branching off. Each unbroken chain of ancestors and descendants is called a lineage. Branches occur when a lineage splits, indicating that a new species has emerged. Scientists infer a lineage split by observing traits that are different from the ancestor species.
 Sometimes there is a gap in phylogenetic trees where scientists think there is probably a common ancestor but it has not been found in living organisms or in fossil records. This gap represents a ghost lineage (represented as F in the example above). The most famous case of ghost lineage is the coelacanth. These lobe-shaped fish have a long fossil record spanning about 300 million years. No coelacanth fossils younger than the Cretaceous period (geological period that lasted from about 145 to 66 million years ago) have ever been discovered, so scientists assumed that coelacanths had been extinct for the past 80 million years.
Sometimes there is a gap in phylogenetic trees where scientists think there is probably a common ancestor but it has not been found in living organisms or in fossil records. This gap represents a ghost lineage (represented as F in the example above). The most famous case of ghost lineage is the coelacanth. These lobe-shaped fish have a long fossil record spanning about 300 million years. No coelacanth fossils younger than the Cretaceous period (geological period that lasted from about 145 to 66 million years ago) have ever been discovered, so scientists assumed that coelacanths had been extinct for the past 80 million years.
However, in 1938 a living coelacanth was caught off the coast of Africa, and a second population was discovered in Indonesia. It turns out that coelacanths had not gone extinct, they just had an 80-million-year ghost lineage. The coelacanth represents a Lazarus taxon, which is a group that reappears after a long period of apparent extinction. Another type of ghost lineage extends a group’s phantom record backward in time, and they are discovered by studying evolutionary relationships like those described above.
How do scientists prove evolutionary relationships without fossil remains?
Several studies have provided evidence that the transfer of genetic information from one species to another, called introgression, can allow extinct species to live on in the genomes of present-day species. In some cases, evolutionary relationships between an extinct lineage and a living lineage are uncovered by sequencing ancient DNA from fossils. For example, using genetic material from fossil remains, scientists have shown that the modern human genome contains genetic traces from at least two extinct hominid relatives: the Neanderthals and the Denisovians. Similarly, modern-day brown bears share some of their genetic sequence with extinct cave bears.
Given that fossilization is a rare phenomenon and that most fossils do not yield sufficient genetic material to sequence, scientists are probably missing numerous ancient genetic transfer events that left traces of extinct species in present-day genomes. So how do scientists prove that these ghost lineages actually existed? By comparing the genomes of living members of the family tree, scientists can look at alterations in genes that arose in each branch as they split off from one another. The presence of genetic variants that do not line up with those already associated with known species could indicate a ghost lineage.
Ghostly lineage of dog
Advancements in DNA sequencing technology have facilitated the sequencing of more organisms than ever before. By combining genetics with evolutionary biology and morphology, scientists are discovering many potential ghost lineages that would have remained hidden without genetics. By lining up the genomes of known living or extinct members of a species, scientists can identify genetic variations that do not show up in the genome of known members, suggesting a potential ghost lineage. 
HudsonAlpha Institute for Biotechnology Faculty Investigator, Faculty Chair and Smith Family Chair for Genomics Greg Barsh, MD, PhD, and senior scientist Chris Kaelin, PhD, along with a team of collaborators, recently uncovered a ghost lineage of dogs while studying dog coat color. The study which was published in Nature Ecology and Evolution explores genetic relationships of a coat color variant called ASIP in modern dogs, wolves, and ancient wild dogs. While performing genomic analysis, the team uncovered a surprising connection between dogs, wolves, and an ancient wolf-like species that is now extinct.
Looking at the different ASIP variants in each of the species suggested a common origin of dominant yellow coats in dogs and white coats in wolves. Using this information, they created phylogenetic trees showing evolutionary relationships between dogs, wolves, and 8 other wolf-like species. The team showed that the dominant yellow allele in dogs and wolves is much older than either species, at least 2 million years, and therefore represents “ghost DNA” from a mysterious wolf-like species.