An Everyday DNA blog article
Written by: Sarah Sharman, PhD
Illustrated by: Cathleen Shaw
Craving a candy bar? No problem! In today’s world, indulging your sweet tooth is a breeze thanks to the abundance of processed sugar. This pantry staple has a fascinating history. Its roots trace back to sugarcane, a tall grass that produces incredibly sweet juice. Scientists and historians think sugarcane was first cultivated in New Guinea as early as 8,000 BC. As a highly valuable commodity, sugarcane cultivation spread like wildfire across continents, becoming a cornerstone of global trade. Today, sugarcane is used for much more than sugar, including bioethanol and other bio-based materials.
Sugarcane thrives in tropical climates but requires significant amounts of fertilization, weeding, and water. Traditional breeding methods created new varieties of sugarcane that can grow in new environments and survive some pathogens; however, genome-directed breeding could help speed up the process of creating new varieties of sugarcane to thrive in our changing world. Let’s learn about sequencing plant genomes, explore the challenges of sequencing complicated plant genomes, and the journey of one research team to sequence the sugarcane genome.

Not all genomes are created equal
Recent advances in genome sequencing have enabled groundbreaking discoveries across many fields. There are large-scale efforts to sequence the genomes of diverse populations of humans through programs like All of Us. There are also massive collaborative projects looking to sequence every organism on the planet, like the Darwin Tree of Life, the Open Green Genome, and more. Despite all of this excitement around large-scale sequencing projects, not all genomes are created equal, and some are much harder than others to sequence and assemble. Many plant genomes are notoriously difficult, but why?
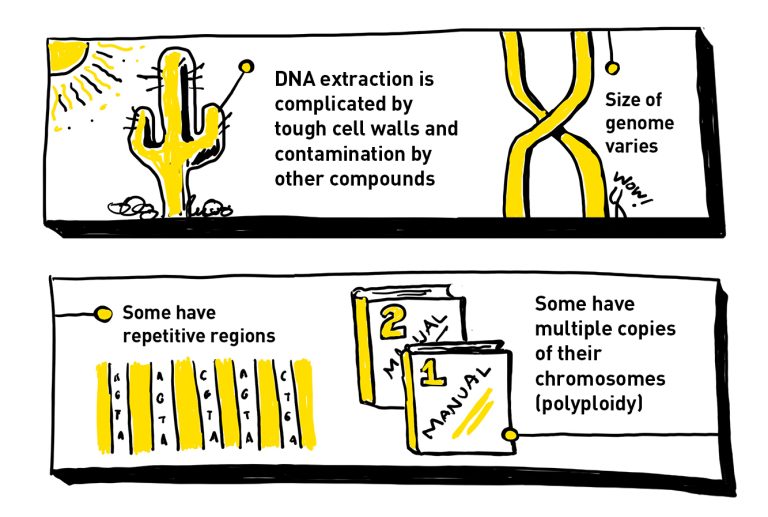
To create a high-quality genome, you need to start with high-quality DNA. Many plant cells have tough cell walls that make it tricky to isolate DNA. They may also contain compounds that can contaminate the DNA sample, throwing a wrench into the sequencing process.
Many plant genomes are enormous, dwarfing the human genome by many times. For example, Paris japonica, a flowering plant, holds the record for the largest sequenced genome at a whopping 149 billion base pairs. In contrast, the human genome is a mere 3 billion base pairs. Giant genomes mean more data to sort through and assemble, making them more time-consuming and complicated than smaller genomes.
In addition to their size complexity, plant genomes are also riddled with repetitive sequences of DNA. This makes the process of assembling the genomes after sequencing very difficult. Think of it like trying to find a specific word in a lengthy song filled with the same chorus over and over. These repetitive regions make it difficult to determine the correct order, like having multiple sections with the same chorus but different verses.
One of the biggest complications for sequencing and assembling plant genomes is that many plants are polyploid, meaning they have multiple copies of chromosomes within each cell. Humans are diploid, meaning they have two copies of each chromosome, one inherited from both parents. Plants, however, can be either diploid or polyploid, with sometimes more than a dozen copies of each chromosome. Having more copies of each chromosome increases the complexity of figuring out which DNA sequence belongs where.
Polyploidy can occur through cellular mishaps in mitosis and meiosis. In meiosis, the cell division that produces gametes (sperm and egg), chromosomes may fail to separate properly. This can result in a gamete containing an entire extra set of chromosomes. If such a gamete fuses with a normal gamete (containing one set of chromosomes), the resulting offspring will have three sets of chromosomes (triploid). Spontaneous doubling of somatic cells can also cause the doubling of meiotic precursor cells, resulting in gametes containing an extra set of chromosomes.
While it makes sequencing more complicated, polyploidy can offer advantages to plants. Polyploidy increases genetic variation, potentially leading to new traits, tolerance to environmental stresses, and larger and more productive plants.
Why are plant genomes so valuable?
If some plant genomes are so complex to sequence and assemble, why do scientists try to do it? Understanding the genetic makeup of plants is vitally important for the health and well-being of all the organisms on our planet.
By decoding a plant’s complete set of genetic instructions, scientists can trace evolutionary relationships between different species, understanding how plants diversified over time. Studying genetic changes in plants over time can reveal plants’ unique adaptations to thrive in various environments, such as drought resistance genes in desert plants or cold tolerance genes in arctic flora. Analyzing plant genomes also helps us pinpoint the genes responsible for essential biological processes in plants, like photosynthesis or disease resistance. For instance, by comparing the genomes of maize varieties with high and low yields, researchers can identify genes that influence grain production, leading to the development of more efficient crops.
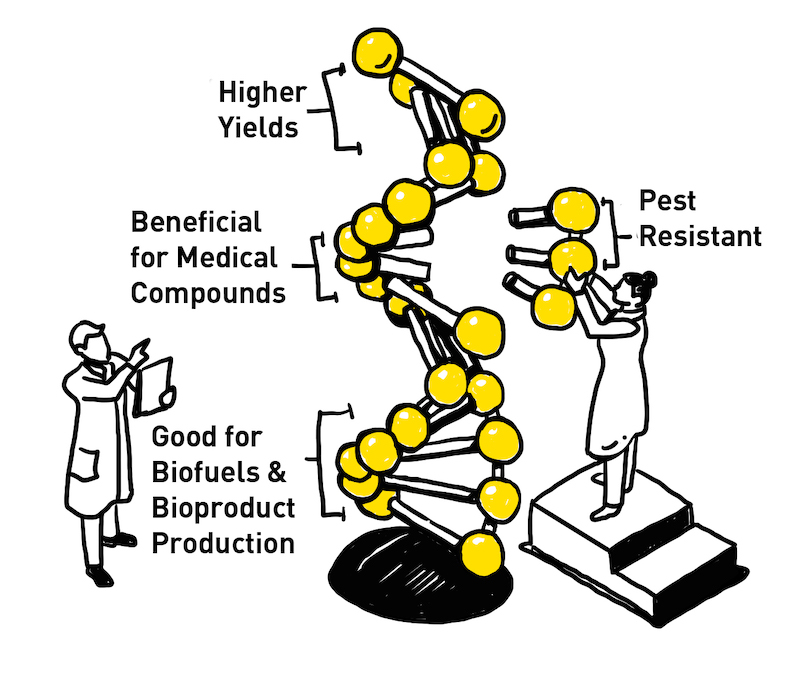
In this way, genetics is poised to help secure our food supply. Information about the genetic makeup of crops allows scientists to develop improved varieties. This can involve breeding plants with higher yields, resistance to pests and diseases, or tolerance to harsh environmental conditions like drought or salinity. Scientists can create crops with improved nutritional value or better storage qualities by manipulating genes.
Plants have long served as a natural source of medicinal compounds. Deciphering plant genomes helps identify genes responsible for the production of these beneficial molecules, leading to the development of new drugs and therapies. Plants can also be engineered to produce biofuels or other industrial products through genetic modification techniques informed by genome sequencing.
Plants play a critical role in maintaining healthy ecosystems. They provide food and habitat for animals, regulate water cycles, and produce the oxygen we breathe. Understanding their genomes empowers us to develop strategies for a more sustainable future, helping to develop strategies for conservation, bioremediation (cleaning up polluted environments), and engineering plants to better capture and store carbon dioxide.
How did scientists sequence the complex sugarcane genome?
Now that we appreciate the difficulties that can arise when sequencing plant genomes and the importance of having high-quality plant genomes let’s return to our sugarcane story. Despite being the most harvested crop by tonnage, sugarcane was one of the only major cash crops without a high-quality genome until recently. Sugarcane has a complicated genome because of its size and the sheer number of chromosomes it has. On average, sugarcane has 12 copies of each chromosome and a total of 114 chromosomes with highly repetitive regions.
When breeding new sugarcane varieties, breeders use only a select number of high-performance parents when making crosses. Because the same parents get used over and over in the breeding process, many of those chromosome copies have large sections that are identical to one another.
Thanks to scientists at the HudsonAlpha Institute for Biotechnology Genome Sequencing Center (GSC) and the Department of Energy Joint Genome Institute, along with collaborators across the globe*, there is now a high-quality sugarcane genome for a type of sugarcane called R570. The GSC have long been experts in sequencing complex plant genomes, but the sugarcane genome clocks in as the most complicated genome they’ve assembled to date.
How did they tackle the monster sugarcane genome? They combined multiple genetic sequencing techniques to obtain the high-quality genome, including long-read sequencing, a newer technique that sequences larger pieces of DNA. The longer DNA sequences make it easier for scientists to find overlapping sections when reassembling the genome. It’s about finding which technique works best for each situation. Polyploidy and the practices breeders use to generate new sugarcane varieties make the genome very complicated to assemble. By combining multiple techniques, scientists were able to leverage each technique’s strengths to overcome this complexity.
Having a high-quality reference genome is a game changer for the sugarcane industry. Using the genome, scientists have already discovered two genes that protect sugarcane from brown rust disease, a notorious foe for sugarcane breeders and farmers. The reference genome will help accelerate sugarcane breeding and the adaptation of sugarcane to our changing environmental conditions.

*This was a massive collaborative project involving many institutions around the world. Significant collaborators include CIRAD, UMR-AGAP, ERCANE, CSIRO Agriculture and Food, Queensland Alliance for Agriculture and Food Innovation/ARC Centre of Excellence for Plant Success in Nature and Agriculture – University of Queensland, Sugar Research Australia, Institute of Experimental Botany of the Czech Academy of Sciences, Corteva Agriscience, Joint BioEnergy Institute, and the Arizona Genomics Institute.
A.L. Healey et al. The complex polyploid genome architecture of sugarcane. Nature, published online March 27, 2024; doi: 10.1038/s41586-024-07231-4



