The HudsonAlpha Software Engineering team both develops and makes use of applications to analyze genomic and clinical datasets to support the interpretation of genomic data. A major interest is in using these tools and methods to provide definitive diagnoses in the case of Mendelian disease. The team provides software and informatics support for the HudsonAlpha Institute for Biotechnology and tools for clinical genomics applications such as those used at the HudsonAlpha Clinical Services Lab, LLC., to help patients and their physicians to find answers to unknown or misdiagnosed diseases.
In addition to clinical application for rare disease, the team is also involved in the application of whole genome sequencing in a research setting to identify causative mutations in individuals with a variety of presumed single gene or complex disorders. HudsonAlpha has a number of ongoing internal and external collaborations aimed at analyzing data from individuals, families, or cohorts to uncover the presumed disease associated molecular changes such as the Clinical Sequencing Exploratory Research program CSER and SouthSeq. Goals are to not only identify variants but also to identify molecular relationships that assist in the selection of new or repurposed treatments.
Classification, Evidence, and Reporting
The team also supports the analysis of genomic sequencing data from the sequencer to the final clinical report converting raw sequencing data into clinically actionable information. Currently supported analyses include DNA sequencing data alignment and variant calling, ChIP analysis, methylation analysis, and RNA-seq analysis, as well as development and application of various methods for data QC, integration, and visualization.
The Software Engineering team develops tools and techniques that fuel scientific progress. We believe that by driving forward the technology of genomics, discoveries will follow — and will impact the world around us at a rate, scale, and scope heretofore unimagined. We recognize that the ability to acquire DNA sequences has grown at an unprecedented rate, and we believe this massive amount of data can be applied with scale and scope not previously appreciated. By testing the limits of sequencing technology, the team has developed unparalleled expertise in the acquisition, analysis, and application of genomic information.
Technology Mission. As a trusted advisor to our Institute, the software engineering team develops innovative solutions to advance the research missions of HudsonAlpha and strives to help deliver ongoing exceptional technology solutions to help bring genomic health to physicians and their patients by producing actionable recommendations and with real-world results.
Software Tools for Genome Interpretation
Sequencing the human genome was the initial obstacle to translating genomic understanding into applications that improve human health and well-being. Today, with next-generation sequencing technology, sequencing is no longer a barrier. The new challenge lies in processing the wealth of data into information that can be applied in the development of new diagnostics, treatments, medicines, materials, or processes.
HudsonAlpha’s team aim to extract biological signal — useful information about how cells, tissues, and organs develop and function — from the billions of points of genomic data generated by HudsonAlpha’s high-throughput sequencers every day. Comprised of biologists, computer scientists, statisticians, mathematicians, and software developers, the computational and informatics teams work creatively and collaboratively to fortify scientific research with value-adding analysis and interpretation. By continuously developing new and novel analysis techniques, they are able to return data with rigorous quality control in a form that has the greatest value to scientists using the data. One of these tools is Codicem.
CODICEM – Extracting knowledge from genomic data
Codicem, referred to as Codi for short, is a clinical tool for the interpretation of whole genome sequencing data. Codi was originally created by the Software Development & Informatics group in Dr. Liz Worthey’s lab and is now developed and maintained by the Institute’s software engineering team. They work hand-in-hand with HudsonAlpha Institute for Biotechnology researchers and clinicians to streamline and expedite genetic variant analysis and interpretation.
The Workflow
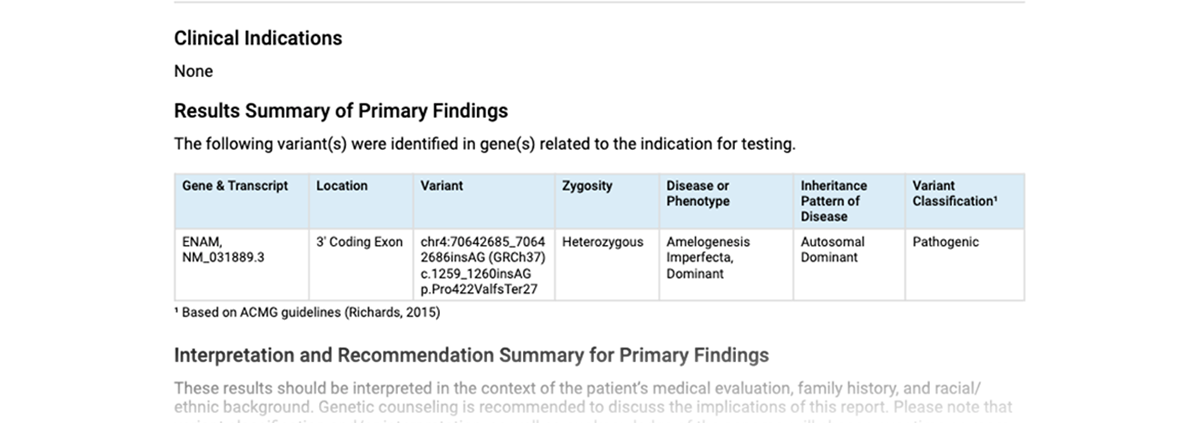
When a patient is referred for clinical whole genome sequencing, a clinical laboratory, like the HudsonAlpha Clinical Services Lab sequences the genome. They then load the genomic variant data into CODI where the data is annotated and displayed to clinicians and researchers.
Most humans have 5-8 million variants in their genome compared to a reference genome, most of which are benign variants. By using a combination of the current knowledge and published data on genetic variants, CODI narrows the millions of variants down to about a couple of hundred variants based upon the user’s requested variant filters. Variant filters can be tailored on the fly by the user.
CODI displays the annotated variant on the computer screen. Researchers and clinicians, alone or in teams, can review the variants and supporting evidence to make a narrower list of potential variants. CODI prepares a report based upon variants of interest chosen by the researchers and clinicians. The report is then signed and returned to the patient’s clinician.
- Patient is referred for clinical whole genome sequencing with interpretation
- Whole genome is sequenced at the HudsonAlpha Clinical Services Lab
- The genomic sequence data is input into CODI software and variants are detected
- Most humans have 5-8 million variants in their genome compared to a reference genome, most of which are benign variants. Rare variant is ~1 in millions.
- CODI filters out variants based on current knowledge and published research on genetic variants
- Narrows down to about 100 potentially relevant variants
- CODI annotates the variants and displays the variants on screen
- Researchers and clinicians (alone or in teams) can review the variants and supporting evidence to make a narrower list
- CODI prepare a report based upon variants of interest is chosen by researchers/clinicians
- The report is returned to the patient’s clinician
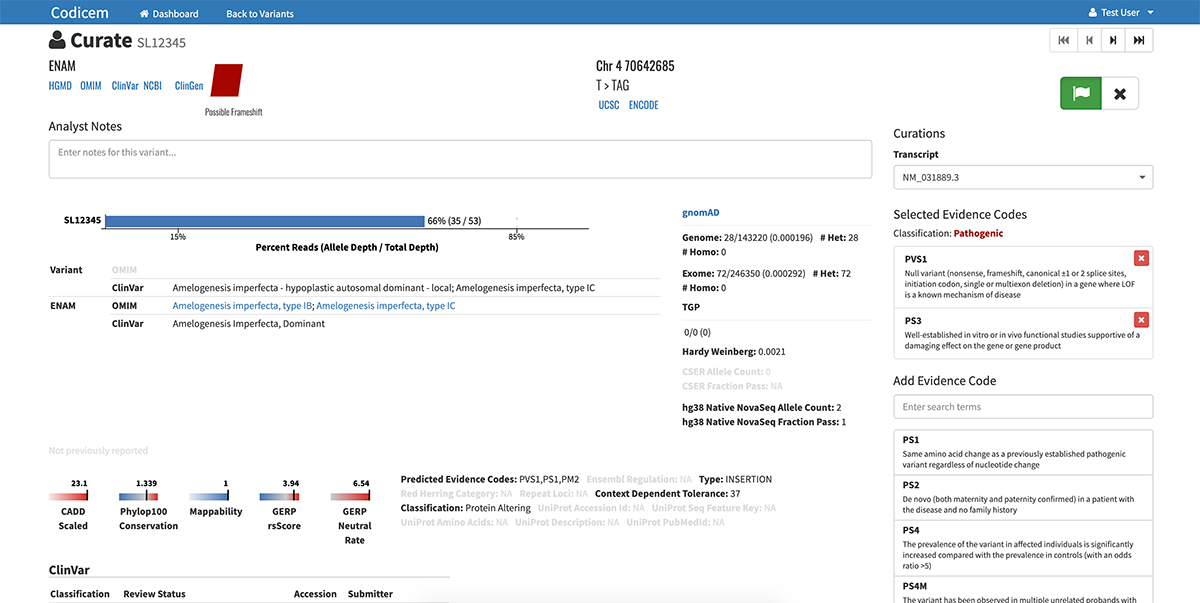
Interactive display of variants
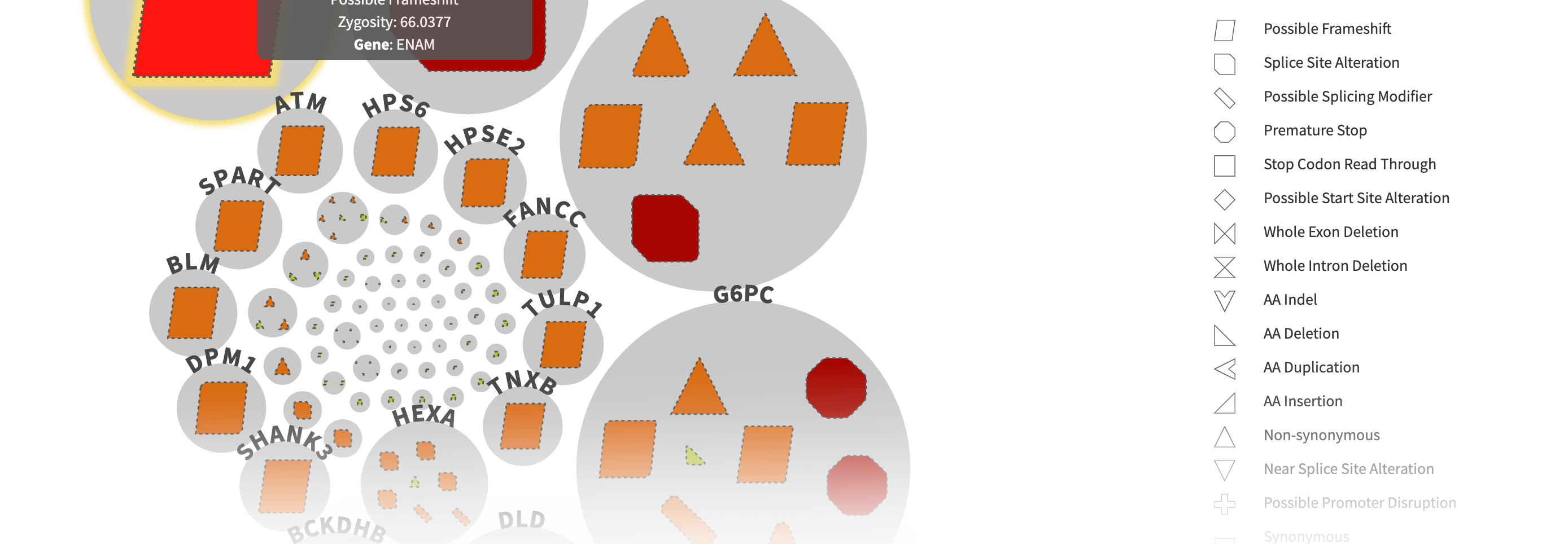
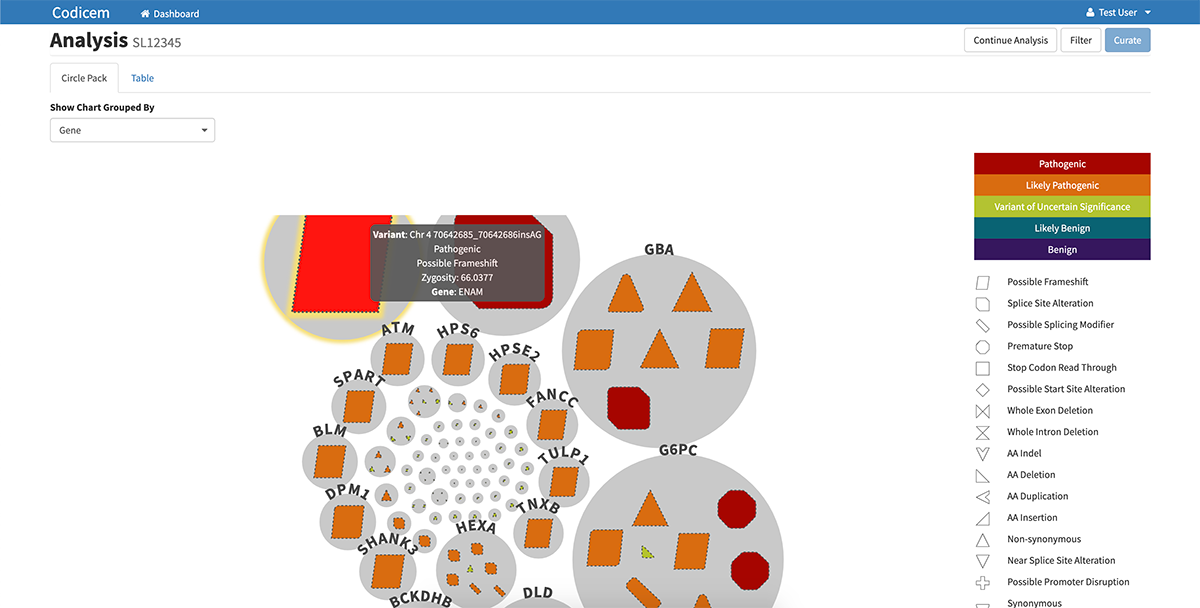
CODI has a user-friendly interface for reviewing variants of interest. After filtering the variants, CODI displays the variants on a computer screen in a visually-engaging, interactive graphic, referred to as a circle pack.
– Each gene in which variants were found are represented by a circle in the circle pack, with the predicted most relevant gene listed first.
– The variants in each gene are embedded within each circle. Different types of variants, like frameshift mutations, indels, etc., are represented by different shapes.
– CODI also labels each variant with a color indicating the likelihood of pathogenicity, ranging from pathogenic to variant of uncertain significance to benign.
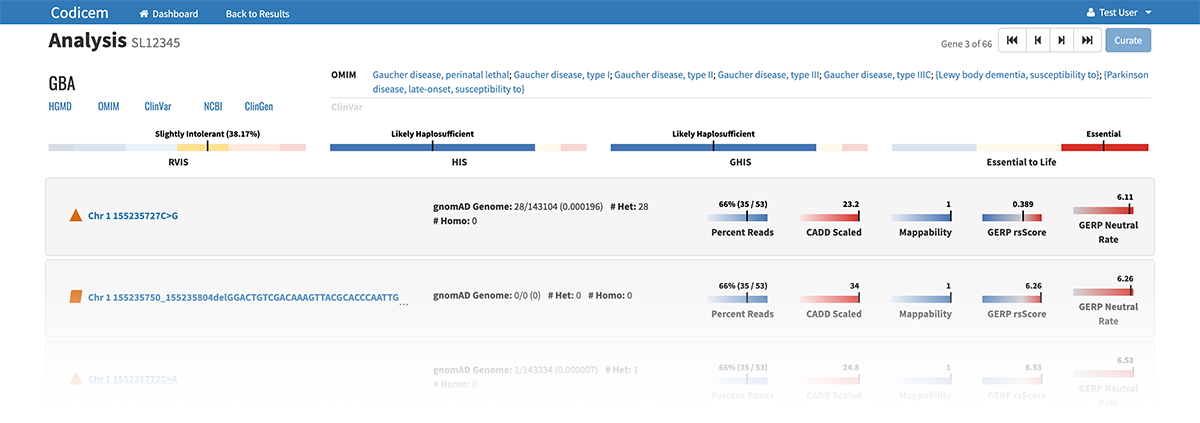
– Clicking on a specific gene circle or variant shape on the circle pack takes the user to a separate page that lists identifying features of the gene or variant, including current information known about diseases related to each genetic variant.
The variants are annotated with levels of evidence according to the American College of Medical Genetics and Genomics (ACMG) guidelines for the interpretation of sequence variants.
The software allows researchers and clinicians to either work alone or in teams to review the list of variants and their supporting evidence. Users also have the ability to input notes about the different variants within the software.
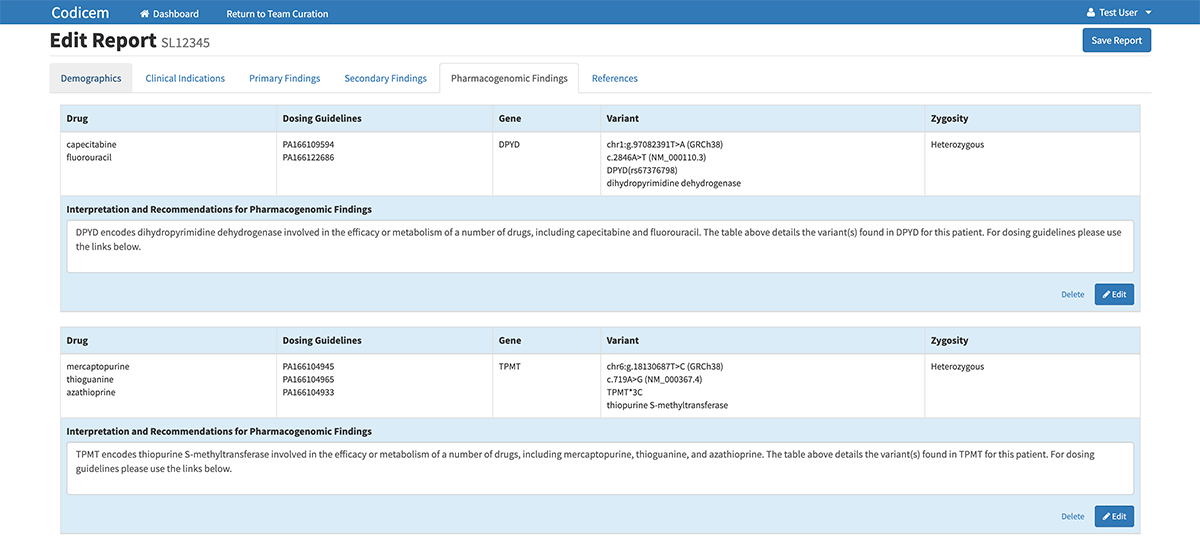
Variant Report
A unique feature of CODI is its ability to generate a detailed variant report. If the individual or team decides a variant is important, they can tell CODI to push the variant into the report. In this way, the report only includes the most potentially relevant variants as determined by the user.
In addition to identifying data about the variant itself, the report also includes a CODI-generated, one-sentence description based on the chosen evidence codes of why the variant may be relevant and important in the disease diagnosis.
The finalized report can be sent to the ordering physician. CODI even has the ability to create tailored reports for different projects or cases.
Uses for CODI Software
The human genome contains about 6.4 billion base pairs, so finding a rare variant is like finding a needle in a haystack. Whole genome sequencing is being used more frequently in rare disease diagnosis to find these one in a million variants. But even after a patient’s whole genome is sequenced, an intensive search for rare variants in the genome still ensues. CODI software was designed to help speed up this diagnostic odyssey This technology can identify mutations that are responsible for rare and common diseases, help patients end their diagnostic odyssey and avoid misdiagnosis, predict the likelihood of future disease development, and help identify which drugs or therapies will work best for individual patients.
At HudsonAlpha, the CODI software technology is being used in the analysis of sequencing data at the HudsonAlpha Clinical Services Lab, and in several research labs. In addition, the physicians at The Smith Family Clinic for Genomic Medicine, LLC., receive CODI-generated reports and use them to help find answers to patients’ undiagnosed or misdiagnosed diseases.
While CODI was designed with whole genome sequencing in mind, the software can also analyze variants in whole exome sequencing and other types of genetic tests. This link shows some of the genomic test available at the HudsonAlpha Clinical Services Lab: https://clinicallab.org/test-menu/. The CODI software also has the ability to annotate pharmacological variants that could help identify which drugs will work best for individual patients.
The HudsonAlpha Software Engineering Team:
Scott Newberry – Director of Software Engineering
Jacob Kelly – Software Developer
Fariba Shaterferdosian – Software Developer
Wayne Schroer – Software Developer
Will Herrington – Software Developer
Contact: For more information about the HudsonAlpha Software Engineering group, contact Scott Newberry at: snewberry@hudsonalpha.org.