An Everyday DNA blog article
A conversation with HudsonAlpha researcher Stephanie Felker, PhD
Written by: Sarah Sharman, PhD
It is widely accepted that many diseases are genetic in nature. An estimated 7,300 Mendelian diseases caused by genetic changes in one gene have been identified. New gene-disease connections are made each year. However, scientists and physicians fail to find a genetic cause in many individuals who are suspected to have a Mendelian disease.
Neurodevelopmental disorders (NDDs) affect 1-2% of children and display a wide phenotypic range that includes seizures, developmental delay, autism, and intellectual disability. These are often Mendelian diseases that result from rare genetic variation. Although scientists have identified nearly 1500 NDD-gene associations, genomic testing in children with NDDs only returns an answer about half the time, suggesting standard analyses may miss some clinically relevant rare variants.
Most genomic analysis looks at parts of the genome that code for proteins. However, emerging data suggest that some non-coding regions might be involved in disease manifestation. Poison exons are one such genomic element recently identified as an important contributor to NDDs. To learn more about poison exons and their implications in neurological diseases, I spoke with Stephanie Felker, PhD, a recent University of Alabama in Huntsville graduate who did her graduate research at HudsonAlpha Institute for Biotechnology in the lab of Greg Cooper, PhD.

Sarah Sharman: First of all, congratulations on your recent successful thesis defense and graduation! The poison exon project was a big part of your graduate school career. Can you set the stage for how you began working on this project?
Stephanie Felker: Thank you! My main research drive is evolutionary biology. A really interesting thing about these poison exons is that they’re very highly conserved in things with a central nervous system. You can find poison exons in many unrelated animals, including ducks, crocodiles, koalas, possums, etc. They’re pretty evolutionarily conserved, which is interesting because they reside in non-coding regions deep within introns. So that is what initially drove my interest.
And because Greg [Cooper] knew that evolutionary conservation and its utilization in genomic analysis was my research inspiration, he invited me to a meeting about a project studying the poison exon in SCN1A. And so I was really fortunate to be pulled into this research literally within my first year at HudsonAlpha. I contributed to a paper and did some fascinating analysis. My primary driver is evolutionary biology, but to make it medically applicable was a goal I never imagined I would reach so early in my career.
Sarah Sharman: Can you describe what a poison exon is? The name sounds pretty scary to me.
Stephanie Felker: The name does sound scary, but actually, we all have poison exons. It’s not like, ‘Oh, you have a poison exon, so you have a disease,’ which is the problem with having the name “poison exon.” Poison exons are alternatively spliced exons that are not typically included in the final mRNA transcript of the gene. In normal biology, DNA bases are transcribed into RNA and mRNA is translated into amino acid chains. When poison exons are included in mRNA transcripts, they prematurely stop the process of translation by either inserting a stop site or causing a frameshift in the DNA that creates a stop site far before the natural one.
Our cells have a way of shutting down the translation of any transcripts that have a stop site in the middle of the transcript. The ribosome will stop on mRNA, so you’ll have a partially translated amino acid chain on one side and a dangling piece of untranslated mRNA on the other. Protein complexes called exon junction complexes at splice sites of the mRNA interact with termination factors and the ribosome, causing the decay of both the transcript and any translated protein. So if these poison exons are included, it causes this early stop event. It causes the transcript to be degraded and no productive protein to be made.
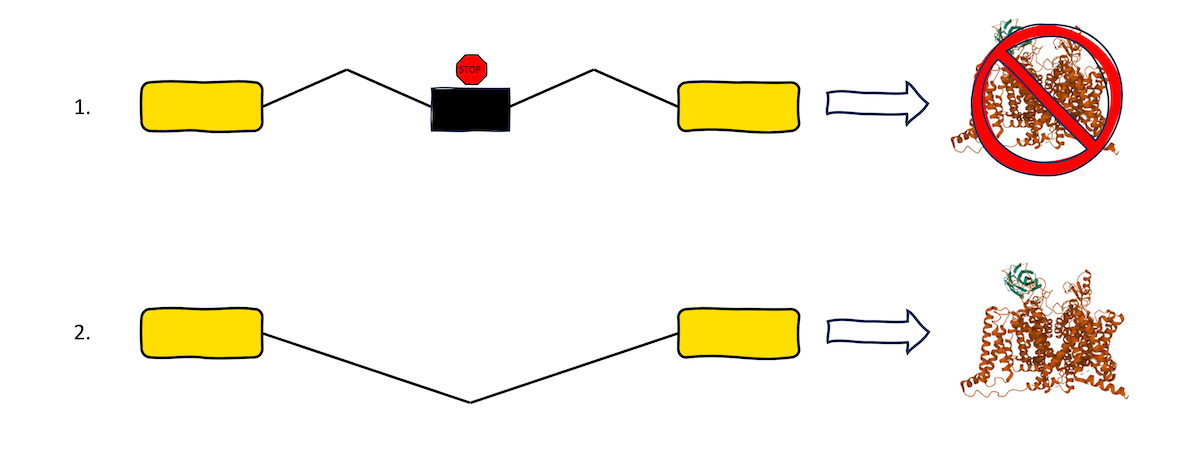
This image shows the different protein fates that occur (1) when a poison exon is included in transcription and (2) when normal splicing is allowed to occur. When poison exons are included, translation stops, and no viable protein product is made.
Sarah Sharman: You mentioned that we all have poison exons, and they aren’t necessarily bad. What do poison exons do in normal biology?
Stephanie Felker: The poison exons I study are involved in regulating the expression of several neurological genes. There are time points in neurodevelopment where you don’t necessarily want your neurons conducting all this signal. So what your cells will do is include these poison exons and say, ‘Okay, let’s not have all of the signal being transduced while we’re migrating or while we’re growing.’ If you don’t have a poison exon variant, you will have certain points of your life where this poison exon is included, depending on what your brain and body need. But for people with the variants, it causes the poison exon to be included all the time. And so you get this loss of function phenotype as you see in Dravet syndrome, like if you have a loss of function SCN1A mutation.
There are poison exons in many different kinds of genes, not just those involved in the brain. Some are involved in protein filamin A, which is the structure of the cell’s cytoskeleton. Others that are fascinating to me are in genes responsible for mRNA splicing. They create this kind of autoregulatory loop–by including this poison exon in the transcript of themselves, they regulate their own transcription. It’s just perfect regulation, like chef’s kiss. So poison exons are all over the place.
Sarah Sharman: Your newest paper, recently published in Genetics in Medicine, asks, ‘Why aren’t poison exons widely included in genome analysis?’ Can you set the stage for why they’ve historically been excluded?
Stephanie Felker: If you look at the genomic analysts in Greg Cooper’s lab–Susan Hiatt, Michelle Thompson, and Don Latner–they are given a proband (individual with a suspected genetic disorder) to look at, and they look at all their genetic variants. If they didn’t filter out variants, they would see thousands and thousands of genetic variants because there’s a great deal of variation in each individual person. There would be alarm fatigue if you included everything, which doesn’t help diagnose patients.
Poison exons specifically haven’t been included in that research because, up until recently, they were not very well characterized. But the widespread use of RNAseq technology allows us to actually look at what the transcript is. Also complicating the matter is the fact that these poison exons are included at very specific points in human development for just a whisper of a time where you can actually capture the transcripts that contain poison exons.
Sarah Sharman: How did your study conclude that poison exons are involved in some diseases and would be beneficial to include in genome analysis?
Stephanie Felker: It had already been established that variants in poison exons can cause neurodevelopmental disorders, but there had yet to be a systematic effort to screen our probands for suspicious variants in introns containing poison exons. I began by looking at mouse data which was helpful because, in mice, you can look at entire developmental time courses, something you can’t do in human datasets. Before molecular studies like this, if it was a seldomly included poison exon, you could determine that the region is conserved across species, but the biological contribution would be largely unknown.
Because this became my thesis project, I had the bandwidth to say, ‘Okay, here are all these hypothetical poison exons. Let’s look across the entire proband dataset Greg Cooper’s lab has assembled to see how many poison exon variants could contribute to disease.’ And because I was just looking at the introns containing poison exons, it was easier than the entire genomic landscape the analysts have to look at. It was a much smaller analytical lift because I was only looking at genes with poison exons, so I could do it in about a year to look at the entire dataset.
I found six poison exon variants across 3000 probands. That is about 0.2 percent, which isn’t a lot. But if you are familiar with our lab’s research and the pie chart we often show during presentations, we only identify variants that are returned to clinical teams in about half of the probands. Adding a little slice for one particular phenomenon is actually significant. I went a step further and determined that if we included these poison exon variants in genome analysis, given the filtration that the analysts already use, we’re talking zero to one additional variants per proband. It doesn’t require deeper sequencing or different technology. It only includes adding these annotations to our current analytical pipeline.
Sarah Sharman: Being able to provide a name for the disease affecting someone or their loved one is meaningful information. Taking it one step further, though, are there treatments that can eliminate the inclusion of these poison exon variants?
Stephanie Felker: Yes, therapeutics are undergoing clinical trials right now that target the 20N poison exon in SCN1A. This therapeutic, called antisense oligonucleotide therapy, increases the expression of SCN1A that does not contain the poison exon, thereby increasing the overall expression of the SCN1A protein. Because of this poison-exon-specific targeting, it is possible that this treatment may work particularly well in probands with poison-exon-related targets. Just another reason why personalized genetic information is so important for many clinicians to inform their treatment plan.
Sarah Sharman: How meaningful was it to be on the cutting edge of a project like this while you were working on your PhD?
Stephanie Felker: It was a dream! When you’re a grad student, it is easy to think that your work is not relevant. But I had confirmation at every point that it was valid work. It was really exciting, and it was a project that Greg was excited about, too, which also helped. I was able to have some opportunities that I wouldn’t necessarily have been able to have if it weren’t “sexy science.” I was given the opportunity to talk at a CESR consortium meeting, which gave me the opportunity to start a collaboration with Dr. Sharon Plon at the Baylor College of Medicine to look at variants causing missplicing in the KidsCanSeq pediatric cancer cohort. I’m immeasurably fortunate for my experiences at HudsonAlpha.


