An Everyday DNA blog article
Written by: Sarah Sharman, PhD, Science writer
Illustrated by: Cathleen Shaw
The ability to sequence and analyze an organism’s DNA revolutionized the field of biology, allowing researchers to understand how the genome functions and how changes in the genome affect all life on earth. Careers in the fields of genetics and genomics are booming. In this new Everyday DNA blog series, Careers in Genomics, we will learn about different career paths in genetics and genomics.
DNA (deoxyribonucleic acid, if we’re being technical) is essential for almost all life on earth. It contains instructions for living things to develop, grow, and function. These instructions are written in a sequence of chemical building blocks called nucleotide bases. There are four bases, adenine (A), thymine (T), cytosine (C), and guanine (G), that combine in different three-base combinations (such as ATG or TGG) to form amino acids, which make up proteins. Scientists discovered more than 20,000 proteins that work together to perform all of the functions in the human body. 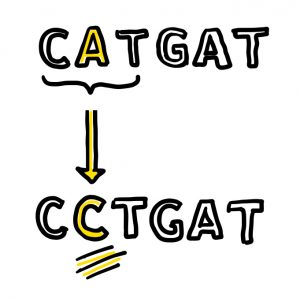
Humans share about 99.9 percent of their DNA with other humans. Although the 0.1 percent difference might seem trivial, it represents about 3,000,000 variants. Some are inconsequential, some cause morphological differences like hair or eye color, and some cause disease. As such, the human genome holds the answers to many of the questions we have about human health and disease.
The ability to sequence DNA and identify variants opened the door to many discoveries related to human diseases and disorders, setting the stage for life-changing tests, treatments, and cures. One field that benefits greatly from genetic sequencing is rare disease research. Let’s learn more about rare disease research and the different professions contributing to the fight to diagnose the undiagnosable.
What is rare disease research?
Rare diseases, by definition, are rare, individually affecting less than 200,000 people in the United States. However, there are currently more than 7,000 identified rare diseases affecting 25 to 30 million people in the U.S. alone. That’s about one in ten Americans. Although their symptoms are wide-ranging, rare diseases often cause chronic illness, disability, and premature death. The cause of some rare diseases like sickle cell, cystic fibrosis, and hemophilia are well understood, and some have successful treatments. However, many rare diseases are still somewhat of a mystery to scientists and physicians.
 Physicians are frequently trained in the diagnosis and treatment of common disorders, but few receive instruction in rare diseases. Because few people have each rare disease, it is common that a patient’s doctor has never encountered another individual with the disease. In other cases, a disease could be so rare that it has not been properly characterized yet. For these reasons and more, individuals with rare diseases often embark on years-long diagnostic journeys, visiting doctor after doctor and often receiving misdiagnoses.
Physicians are frequently trained in the diagnosis and treatment of common disorders, but few receive instruction in rare diseases. Because few people have each rare disease, it is common that a patient’s doctor has never encountered another individual with the disease. In other cases, a disease could be so rare that it has not been properly characterized yet. For these reasons and more, individuals with rare diseases often embark on years-long diagnostic journeys, visiting doctor after doctor and often receiving misdiagnoses.
Most patients suffering from a rare or undiagnosed disease take several medications or interventions to treat their symptoms. An accurate diagnosis opens the door to better treatment strategies, identifying potential new therapeutics, and avoiding unnecessary treatments that could have severe side effects. Advances in genomic sequencing technology help patients avoid a long and painful path to diagnosis.
An estimated 80 percent of rare diseases have a genetic origin, so studying a person’s genome can increase the potential of identifying variants responsible for their symptoms. Clinical genome sequencing looks at a person’s entire genome. Experts can study the bases in the patient’s genome to identify variants responsible for their disease. By identifying the specific DNA changes that cause the disease, clinicians can provide the patient with answers to their long quest for a name for their symptoms.
Diagnosis, treatment, and prevention of rare diseases are only now making some progress. There are still many diseases that researchers haven’t identified a genetic cause for. Rare disease research is paramount to identifying the causes so that more patients are afforded diagnoses. Improving sequencing technology and computational analysis platforms are also important in rare disease research so that scientists can continue to uncover tricky genetic variants responsible for disease.
Who’s who in rare disease research: research scientist 
There is a whole cast of characters who make rare disease research possible. From identifying new gene variants involved in diseases to implementing those findings in a clinical setting, there is plenty of day-to-day work that goes into these monumental discoveries. Many professions are involved in rare disease research, including scientists, computational biologists, geneticists, genetic counselors, physicians, and project coordinators. In this blog post, we are focusing on scientists, but stay tuned for more installments of this series that will focus on other professions.
Research scientists are at the heart of rare disease research, identifying new gene-disease relationships, diving into the molecular underpinnings of the disease, and discovering new treatments. These scientists work at research universities, institutions, hospitals, government agencies, and pharmaceutical companies.
In most research labs, there is a hierarchy of different levels of scientists working together. Often, an experienced scientist called a principal investigator (PI) is at the head of the lab. Scientists at this level typically have a doctoral degree and several years of research experience under their belt. They assume full responsibility for a research project, secure funding for their lab, and mentor other individuals in their lab. PIs are generally more senior scientists with experience performing research and solving complex scientific problems.
Senior scientists plan and lead experiments within the lab. Because they have several years of experience, they operate with little oversight from the PI. Some senior scientists even mentor more junior scientists or students in the lab. In addition to planning and executing experiments, senior scientists also write grant proposals to secure funding for research projects, present their research findings to interested groups, analyze experimental data, write and publish their research in academic journals, and collaborate with other scientists.
Undergraduate and graduate students, technicians, and interns do much of the hands-on work in a research lab. Gaining laboratory experience is a critical part of training to become a scientist.
 There are many different roads to becoming a scientist in the rare disease field. Scientists typically begin their postsecondary educational journey by obtaining a bachelor’s degree in biology, biomedical science, chemistry, genetics, or another related scientific field. A comprehensive understanding of biological processes and human genetics is important for scientists to ask relevant questions and seek the answers in the genome. Technicians do not typically need additional education beyond an undergraduate degree and receive much of their training on the job. Senior scientists and PIs typically require additional education and training. Master’s and doctoral degrees are common tracks for those individuals interested in higher-level science research.
There are many different roads to becoming a scientist in the rare disease field. Scientists typically begin their postsecondary educational journey by obtaining a bachelor’s degree in biology, biomedical science, chemistry, genetics, or another related scientific field. A comprehensive understanding of biological processes and human genetics is important for scientists to ask relevant questions and seek the answers in the genome. Technicians do not typically need additional education beyond an undergraduate degree and receive much of their training on the job. Senior scientists and PIs typically require additional education and training. Master’s and doctoral degrees are common tracks for those individuals interested in higher-level science research.
Computational biologists and data scientists are becoming increasingly important in rare disease research. As scientists deal with ever-growing genome sequencing datasets, they need better ways to handle and analyze the data to extract the information they need to diagnose diseases. Computational biologists and data scientists are trained to use computational solutions to extract meaning from DNA sequences composed of billions of base pairs. To learn more about careers in computational biology, read the first installment of this blog series, “Careers in Genomics: Computational biologist.”
Spotlight: Greg Cooper lab at HudsonAlpha
Many scientists and genetic counselors at the HudsonAlpha Institute for Biotechnology are passionate about identifying and characterizing genes involved in rare developmental disorders and translating the findings into clinical applications and disease diagnosis. HudsonAlpha Faculty Investigator Greg Cooper, PhD, and his lab use genome sequencing technology, along with experimental and computational techniques, to identify disease-causing genetic variants within an individual’s genome.
The lab is part of several research studies that enroll children with rare and undiagnosed diseases and provide them with DNA sequencing. Cooper’s lab and their collaborators have enrolled and sequenced the genomes of more than 1,800 children and sometimes their parents. The group found the genetic cause for about 30 percent of affected children through the CSER1, SouthSeq, and AGHI studies.
Sometimes, detecting a variant is not enough. Many of those detected during genome sequencing do not have enough evidence to prove they cause disease. A cohort of many individuals with overlapping symptoms and variants in the same gene is important for proving a causal relationship between the gene and the disease. However, because so few individuals have any rare disease, there could be only one case in a given state, region, or even country. A freely accessible website called GeneMatcher allows scientists and geneticists to collaborate and create larger cohorts of patients. The scientists input their gene/disease discoveries to the site and match with others who made similar findings. The Cooper lab submitted 280 genes to GeneMatcher as of late 2022, leading to 25 collaborative publications linking dozens of genes to developmental disorders, including their newest discovery: a gene called ZMYM3 linked to neurodevelopmental disorders.
What about the 70 percent of individuals who did not find a diagnosis? The team believes that many neurodevelopmental and neuromuscular disorders result from genetic variation that cannot be detected using standard short-read sequencing technology. Cooper and his lab are resequencing the genomes of individuals with neuromuscular disease phenotypes from their research projects that did not receive a diagnosis with standard short-read technology. They are using a newer technology called long-read sequencing that they hope will detect more, oftentimes complex, genetic variation than short-read sequencing. The lab showed its utility already by helping physicians make diagnoses for two pediatric patients with undiagnosed neurodevelopmental disorders.


