An Everyday DNA blog article
Part 2: Next-generation sequencing
Written by: Sarah Sharman, PhD, Science writer
Illustrated by: Cathleen Shaw
Part 1 of our blog series on The Evolution of DNA Sequencing Technology described the early days of DNA sequencing and brought us through the completion of the Human Genome Project in 2003. Scientists were excited by the completion of the first human genome—not just because it was a monumental accomplishment, but because it showed what was truly possible with DNA sequencing technology and collaborative science.
However, the efforts to sequence larger genomes made the need for cheaper, large scale and high efficiency sequencing methods apparent. In Part 2 of this blog series, we will look into the transition from automated Sanger sequencing to massively-parallel, next-generation sequencing.
Amplifying our way to larger genomes
The completion of the Human Genome Project ushered in a new era of rapid, affordable and accurate genome sequencing called next generation sequencing (NGS). While several different NGS platforms have been developed in the past two decades, they all have a common feature: sequencing many different DNA fragments at once by running millions of reactions at the same time.
How did scientists move from performing one reaction on one DNA fragment at a time to running many reactions on many DNA fragments at once? The answer is a technique called DNA amplification. Similar to Sanger sequencing, scientists break the DNA being sequenced into smaller fragments. However, instead of having to be inserted into a bacterial vector and copied before sequencing, the fragments are attached to a solid surface like a microscope slide, microchip or a nanobead and amplified, or copied, creating clusters of DNA, each originating from a single fragment. Then many sequencing reactions can be run on each cluster simultaneously.
The major differences between NGS platforms are mainly in the type of DNA sequencing that each uses on the clusters of DNA. Pyrosequencing, developed in 1996, was the first alternative to the conventional Sanger sequencing method. During pyrosequencing, bases are added one at a time to the reaction and if the correct base attaches to the template DNA strand, it releases a chemical called pyrophosphate which produces light that is measured by a camera. Interestingly, luciferase, the enzyme that converts the pyrophosphate to light, is the same enzyme that makes fireflies glow.
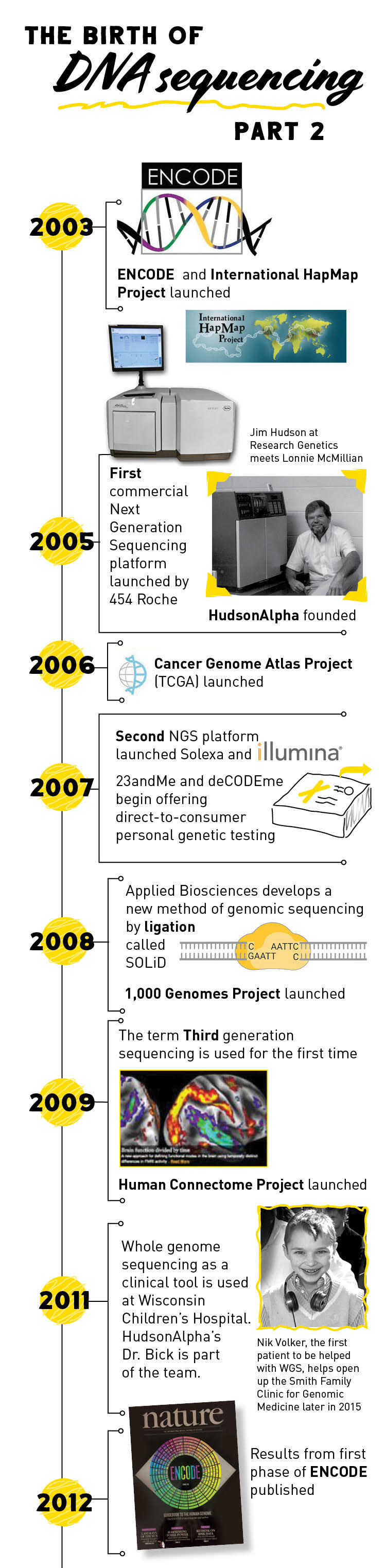
The principles of pyrosequencing laid the groundwork for the development of the first high-throughput, next-generation sequencing platform launched in 2005 by 454 Life Sciences (later acquired by Roche). One drawback of pyrosequencing is that it is hard to tell if the light emitted is for one or more than one of the same base since many can be added at once.
Another company, Solexa (which is now owned by Illumina), introduced a second next-generation sequencing platform in 2007 that used a slightly different sequencing method. In this method, dye-labeled nucleotides are added to the reaction. When a nucleotide binds to the DNA template, it produces fluorescent light. The dye-labeled nucleotides also have a terminator so that only one nucleotide can be added at a time, increasing sequencing accuracy.
2008 saw the introduction of a third NGS sequencing platform called SOLiD (Supported Oligonucleotide Ligation and Detection). SOLiD uses short, dye-labeled DNA probes to generate a DNA sequence. If the two nucleotides on the probe match the next two nucleotides in the template strand, they attach to the template and produce a fluorescent signal.
No matter which NGS platform you use, they all offer many advantages over Sanger sequencing. NGS is significantly cheaper and faster than Sanger sequencing. In addition, NGS requires significantly less DNA to obtain a sequence. Sanger sequencing requires several strands of template DNA for each base being sequenced—for a 100 base pair sequence you would need many hundreds of copies of the template DNA.
Beyond the genome: epigenome, transcription factors and more
Scientists immediately started taking advantage of the rapid advances in DNA sequencing technology to begin making sense of what was discovered in the Human Genome Project. Remember in Part 1 we learned that humans have about 3.1 billion nucleotides that make up about 20,000-25,000 human protein-coding genes. It turns out that these protein-coding genes only make up a fraction of our genome, leaving the function of most of the genome a mystery.
In the years following the conclusion of the Human Genome Project, several other large, collaborative projects were initiated to start unraveling the genome. Both the Encyclopedia of DNA Elements (ENCODE) Project and the International HapMap Project began in 2003 right on the heels of the Human Genome Project. ENCODE, which is still ongoing today, seeks to identify all of the functional parts of the human genome (To learn about HudsonAlpha’s contributions to ENCODE, see Research Highlight #2).
The International HapMap Project, which published its final dataset in 2009, was created to identify common genetic variation among humans. The 1000 Genomes Project, launched in January 2008, continued the work of HapMap. It was the first project to sequence the genomes of a large number of people, reporting final analysis on the genomes of more than 2,500 individuals.
The Cancer Genome Atlas began in 2005 and looked specifically at genetic mutations responsible for cancer, bringing together genome sequencing and bioinformatics. The project, which brought together scientists from diverse disciplines and multiple institutions, characterized over 20,000 primary cancer and matched normal samples spanning 33 cancer types.
Like the Human Genome Project before them, information produced by ENCODE, HapMap, Cancer Genome Atlas and 1000 Genome Project were made freely available to the worldwide scientific community through public databases.
Next-generation sequencing in the clinic
Advances in genomic sequencing technology and the open sharing of data sparked a genomics revolution that led to countless discoveries in the biological sciences and beyond. Scientists could now link diseases and other conditions to underlying genetic causes.
In the decades since the completion of the Human Genome Project, whole genome sequencing technology has advanced and become more cost effective. As such, the technology is being applied more readily in basic and clinical research settings.
Projects like the 1000 Genomes Project began sequencing many human genomes to pinpoint disease causing variation in the human genome. It was not until 2010 that the first genome containing a disease-causing mutation was sequenced. Scientists found a single-gene variant that causes Charcot-Marie-Tooth neuropathy. This new information proved that it was possible to pinpoint genes associated with certain diseases using whole genome sequences.
As more and more genes associated with disease were discovered, sequencing began to move into clinical diagnostics. In 2011, a team of physicians at Wisconsin Children’s Hospital were among the first to use whole genome sequencing to diagnose and treat a rare disease in a four-year old boy named Nicholas Volker. Information gained from his genome allowed his physician to determine the cause for his nearly fatal, severe intestinal symptoms. Once they figured out the cause of his intestinal problems, Nick received a bone marrow transplant that cured him.
Nicholas’ success story, coupled with many other diagnoses in subsequent years, led to the emergence of genomic medicine, a field of medicine that uses a person’s DNA to guide healthcare decisions and anticipate, diagnose, and manage disease. The information carried within your genome can also help a geneticist identify disease risk factors and guide preventative healthcare decisions, identify disease carrier status, and even identify genetic factors that may have an effect on your reactions to certain drugs.
To learn about the newest stage of genome sequencing technology and its applications to the fields of health and biotech, stay tuned for Part 3 of this series.

ENCODE Project
Although a near-complete human genome sequence was published in 2003, it turns out that knowing the precise sequence of the genetic code is not the end of the story. The Human Genome Project determined the location of over 20,000 genes. However, the areas of the genome that we recognize as genes only take up a small part of the genome, leaving the function of most of the genome a mystery.
The Encyclopedia of DNA Elements (ENCODE) Project began after the Human Genome Project with the goal of figuring out which base pairs are important and what their function is. Hundreds of scientists in dozens of labs across the world were funded to identify these active parts of the genome, like genes that make protein and RNA, genes that turn other genes on and off, and genes that can change a cell’s function.
At HudsonAlpha, Richard M. Myers’ lab, along with the labs of Sara Cooper, PhD and Adjunct Faculty member Eric Mendenhall, PhD, have been major contributors to the ENCODE Project since its inception. Most recently his lab has been studying the on/off switches that control gene expression, called transcription factors. As part of the ENCODE 3 Project, the HudsonAlpha team and their collaborators performed the largest study of transcription factors expressed at physiological, or normal, levels to date.



