Can environmental DNA help protect Earth’s biodiversity?
 DNA — it’s all around us. While it is easy to think that DNA is only within cells, it is actually floating around in the air we breathe, the water we swim in, and the ground upon which we walk. All you true crime buffs know that humans are constantly leaving behind DNA in the form of strands of hair, fingerprints, and even blood. Analyzing the DNA in these trace materials can help identify the person that left them behind.
DNA — it’s all around us. While it is easy to think that DNA is only within cells, it is actually floating around in the air we breathe, the water we swim in, and the ground upon which we walk. All you true crime buffs know that humans are constantly leaving behind DNA in the form of strands of hair, fingerprints, and even blood. Analyzing the DNA in these trace materials can help identify the person that left them behind.
Animals also naturally shed DNA into their environment through sloughed off skin cells, shed hairs, and even in fecal matter. Scientists have recently begun using this environmental DNA to identify animals for a number of fascinating applications ranging from wildlife conservation to pathogen monitoring in aquatic environments.
What is eDNA and how is it detected and measured?
Environmental DNA (eDNA) refers to trace amounts of DNA gathered indirectly from environmental samples rather than directly from an organism. It originates from cellular material shed by organisms into aquatic or terrestrial environments that can be sampled and monitored using laboratory techniques. eDNA has been successfully detected and measured in water, ice cores, soil, sediment, and even the air.
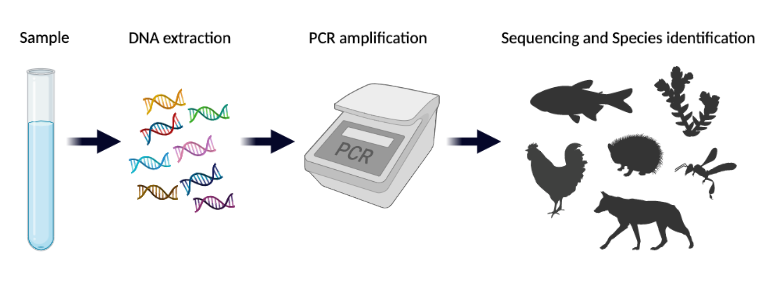
Analyzing eDNA starts with collecting the DNA in an environmental sample. Common environmental samples include soil, water, or air. However, eDNA can also be collected from other materials like the gut or feces of an animal, the petals of a flower, or the bark of a tree, for example.
There are two main approaches for analyzing eDNA recovered from an environmental sample. Most often, eDNA is used to detect the presence of individual species of interest like an invasive or endangered species. Every organism has a unique DNA sequence associated with it.
Scientists can use a technique called quantitative polymerase chain reaction (qPCR) to amplify short stretches of DNA that are unique to the target species. If these DNA stretches are detected, eDNA from the target species is present in the sample, but if there is none detected, eDNA from the target species is not present.
The second method, called eDNA metabarcoding, simultaneously detects many species in the same test from the same sample. In this way, a single sample can provide a snapshot of an entire community of organisms. Metabarcoding begins with PCR amplification of DNA like in the other approach but this is where the similarities end.
After amplification, target genes specific to the species being studied are amplified from the eDNA mixture. The amplified genes are combined into a pool that is compared to a reference library that contains many different sequences derived from known species. If a sequence obtained from the eDNA sample matches that from a known species in the reference library, it indicates that the species’ DNA was present in the original sample.
Practical uses of eDNA
Conservation efforts to protect our planet’s biodiversity depend on the monitoring of species and populations to obtain reliable distribution patterns and population size estimates within specific ecosystems. Current monitoring techniques include animal track identification and monitoring, camera traps, and aerial monitoring. These methods can be costly, time consuming, and do not usually account for rare, shy animals that do not venture into open spaces.
eDNA is a powerful tool for biodiversity inventory and monitoring. It allows scientists to build up a far more detailed image of which species live in the environment. It also enables them to find and identify small invertebrates and microorganisms that live throughout all environments, but are very rarely counted due to their small size.
In addition, eDNA sampling is useful for the early detection of invasive species, especially in aquatic environments. By periodically collecting and sequencing water samples, scientists can detect new and potentially invasive species earlier and more effectively than by sight tracking alone. This method is being successfully employed by the National Park Service in collaboration with the US Geological Survey Alaska Science Center to develop several metabarcoding tests aimed at assessing aquatic communities, including the detection of invasive species.
 Soil and ice are also fruitful materials from which to extract eDNA. DNA fragments shed from animals can persist in soil and ice for thousands of years, meaning eDNA can be used to paint a portrait of ancient ecosystems. Several recent publications have recovered eDNA from the animal and human inhabitants of early communities and cave dwellings. One cave site in Spain contained Neanderthal nuclear and mitochondrial DNA from multiple soil layers deposited 80,000 to 113,000 years ago.
Soil and ice are also fruitful materials from which to extract eDNA. DNA fragments shed from animals can persist in soil and ice for thousands of years, meaning eDNA can be used to paint a portrait of ancient ecosystems. Several recent publications have recovered eDNA from the animal and human inhabitants of early communities and cave dwellings. One cave site in Spain contained Neanderthal nuclear and mitochondrial DNA from multiple soil layers deposited 80,000 to 113,000 years ago.
While detecting eDNA in aquatic environments and soil are being used more frequently, collecting eDNA from the air is not as established. As a proof of concept for using eDNA from the air to detect nearby animals, two groups vacuumed DNA from the air around zoos. Zoos are a useful location because they generally contain animals not found in the surrounding countryside. In each case, DNA was consistently identified from dozens of zoo animals. It’s unclear how much DNA floats off organisms into the air, how long those molecules remain aloft and how far they can travel. Those questions require further study.
Although eDNA monitoring is still a relatively new technique, it is already proving to be useful to the scientific community. Maybe one day there will be little air vacuums monitoring organisms in ecosystems throughout the world.
To schedule a media interview with Dr. Neil Lamb or to invite him to speak at an event or conference, please contact Lara Burhenn by email at lburhenn@hudsonalpha.org or by phone: Office (256) 327-5216 | Cell (256) 937-8210