February 10, 2021 (Huntsville, Ala.) – Scientists at the HudsonAlpha Institute for Biotechnology have a long and fruitful history of sequencing complex genomes, dating back to the Human Genome Project. Although the completion of the first human genome was a monumental accomplishment that sparked the genomics revolution, the human genome is relatively uncomplicated compared to other species. Since the Human Genome Project, the scientists at HudsonAlpha have become experts at sequencing some of the most complicated genomes—plants.
While humans have two copies of every chromosome, some plants have four, six, eight, or even ten or more copies of each chromosome. Complicating their genomes even more, plant genomes are highly variable, having more variants per chromosome pair than the human genome. Plant genomes are also very repetitive which makes assembling their genomes even harder.
Using the ever-advancing next-generation and third-generation sequencing technologies, the HudsonAlpha Genome Sequencing Center (GSC), led by HudsonAlpha Faculty Investigators Jane Grimwood, PhD and Jeremy Schmutz, are masters at generating complex plant genomes. As of early 2021, they have sequenced reference genomes for more than 175 plants—approximately half of the plants sequenced as high-quality references worldwide.
Because of their plant genome expertise, Grimwood and Schmutz’s team is part of a multinational team of researchers that have been studying the genome of a plant called switchgrass for over a decade. New results from their efforts were published in the journal Nature in January 2021, and describe groundbreaking findings that would not have been possible without the presence of a high-quality genome assembly.
A promising bioenergy crop
 Switchgrass (Panicum virgatum) is a native North American plant with widespread distribution across the entire eastern side of the United States, spanning from the east coast to the Rocky Mountains, and from Canada down into southern Mexico. The Department of Energy (DOE) designated switchgrass a promising candidate for biofuel, renewable fuel that is produced from the biomass of plants. As a perennial plant, switchgrass can be harvested for biomass for many years after it is initially planted. It grows on marginal land with little resource input so it will not take land and resources away from food crop production, adding to its promise as a biofuel candidate.
Switchgrass (Panicum virgatum) is a native North American plant with widespread distribution across the entire eastern side of the United States, spanning from the east coast to the Rocky Mountains, and from Canada down into southern Mexico. The Department of Energy (DOE) designated switchgrass a promising candidate for biofuel, renewable fuel that is produced from the biomass of plants. As a perennial plant, switchgrass can be harvested for biomass for many years after it is initially planted. It grows on marginal land with little resource input so it will not take land and resources away from food crop production, adding to its promise as a biofuel candidate.
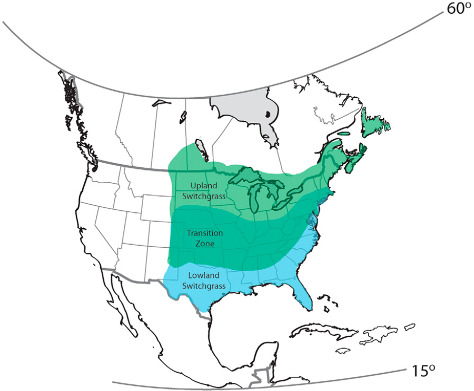
Image reproduced from Casler, M.D., et al. (2011), The Switchgrass Genome: Tools and Strategies. The Plant Genome, 4:. https://doi.org/10.3835/plantgenome2011.10.0026
Regional differences have been observed in switchgrass plants across the country— plants from the northern US, called upland, are often smaller in size compared to plants from the southern US, called lowland, which are large and produce a lot of biomass. However, the southern plants often cannot survive cold conditions in the north. Scientists and the DOE are interested in transposing these traits to create plants that can survive across a range of environments while still producing a large amount of biomass that can be turned into biofuel.
“In order to determine the genetic regions responsible for useful traits in switchgrass, researchers needed a reference point from which to identify differences between the varieties of switchgrass,” says Grimwood. “Our group sequenced the first version of the switchgrass genome in 2008, shortly after moving to HudsonAlpha. Then Jeremy met Tom Juenger at the first switchgrass meeting in 2012, and the collaboration between our genomics expertise and his switchgrass expertise was born.”
The road to the reference genome
Generating the current version of the switchgrass genome was a more than a decade-long project that involved long-term effort from many groups led by researchers at HudsonAlpha, the University of Texas (UT) at Austin, and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab).
Sequencing the switchgrass genome is complicated by the fact that it is a tetraploid , having four copies of its nine chromosomes. More specifically, it is an allotetraploid, meaning two different diploid species came together to produce a single-lineage of switchgrass that kept all of the copies of the parental genes. The four sets of every gene in the genome are split into two subgenomes that come from each of the parental species.
The HudsonAlpha team, led by GSC Genome Analysis Group Leader Jerry Jenkins, PhD, took advantage of advances in long-read DNA sequencing technology to overcome these complications and create a well-resolved, chromosome-scale assembly that splits the two subgenomes, allowing researchers to more effectively pinpoint the locations of genes in each subgenome.
This current version of the switchgrass genome (version 5) is a culmination of over a decade of sequencing and resequencing. According to Schmutz, the completion of the switchgrass genome assembly was complicated, albeit improved, by the ever-evolving sequencing technology. It took almost ten years to complete because the technology kept improving and the team kept updating the genome assembly with the newer technology.
“We sequenced version 1 using Roche 454 sequencing technology, versions 2-4 took advantage of Illumina sequencing, and version 5 rounded it out using PacBio long-read sequencing,” says Schmutz. “We did not start over each time, instead we replaced parts of the genome with the new resolved pieces. Each new technology added clarity to the sequence, allowing us to improve the assembly from 550,000 contiguous sequences of DNA in version 1 to 1,000 in the current version of the genome.”
The high-quality switchgrass genome allows the team to make discoveries that just would not have been possible without it. Until very recently, when researchers wanted to study traits in polyploid crops that have complex genomes like switchgrass, they had to use less complex crop genomes as a model and then infer the trait back into the complex genome. In switchgrass for example, researchers used a plant called Panicum halii.
“The complexity of plant genomes has been a major barrier to developing genetic resources to accelerate effective molecular breeding,” says John Lovell, PhD, HudsonAlpha senior scientist and first author of the manuscript. “Genetic models are useful for gaining a foundational understanding of biology. However, to actually accelerate breeding, we need to find genetic variants that are associated with yield in crop species. Now we can develop genome resources for nearly any species, allowing us to study them directly without the need for a less complex model.”
When high-quality genomics lead to monumental findings
While Schmutz and Lovell were chipping away on the switchgrass genome, collaborators in Tom Juenger’s lab at UT Austin were on a cross-country mission collecting switchgrass from all over its growing region. The team then planted these diversity sets at more than ten different research gardens ranging eight states and 1,100 miles. Using the new version of the switchgrass genome, the research team analyzed the genomes of 732 diverse switchgrass plants from these research gardens to begin mapping out local switchgrass adaptations and linking the traits to their underlying genetics.The team assigned the plants from the common gardens to different populations called ecotypes which are based on visual traits like size and abundance. The switchgrass was grouped into upland, coastal lowland, and Texas lowland varieties (pictured below), and they were also grouped genetically into Gulf (mainly located in the southern US), Atlantic (along the East Coast), and Midwest populations.
When the team started comparing the ecotype groupings with the genetic groupings, things got interesting. As they went up the East Coast, the plants got smaller and hardier, suggesting a transition from one ecotype to another, yet the plants were not genetically much different from one another. Adding to the unusual finding, during a cold winter in 2018 some of these plants that were located in Chicago survived even though their genetic material is lowland, which do not typically tolerate cold. The team reasoned that there must be a small subset of the genome that is strongly contributing to the cold tolerance trait, allowing these genetically lowland plants to survive a harsh winter.

A switchgrass common garden (left) and comparative images of the three ecotypes of switchgrass (right). Images modified from original photos by Robert Goodwin at MSU (left) and Jason Bonnette at UT Austin.
After diving further into the genome, the team found pieces of the genome from a Midwest subpopulation in the Atlantic subpopulation, suggesting that they obtained genes for winter survival from the Midwest subpopulation possibly during the last glacial maxima when the populations would have had the ability to exchange genetic material. This knowledge, combined with the new reference genome, allows the team to map regions of the switchgrass genome that are associated with climate adaptation and fitness.
“Using the last version of the genome, version 4, we simply couldn’t resolve regions of interest,” says Lovell. “The newest version of the switchgrass genome allows us to spot regions in the genome that are associated with important traits like cold tolerance. Once we identify these regions, breeders can use them to develop new strains of switchgrass.”
Researchers from the University of California (UC), Berkeley, Rutgers University, USDA-ARS, Arizona Genomics Institute, University of Georgia, Athens, Clemson University, Marshall University, Jawaharlal Nehru University (India), Noble Research Institute, University of Nebraska, Lincoln, South Dakota State University, University of Missouri, Argonne National Laboratory, USDA-NRCS, Texas A&M University, UC Davis, Oklahoma State University, University of Oklahoma, and Washington State University were also involved in this work.
About HudsonAlpha: HudsonAlpha Institute for Biotechnology is a nonprofit institute dedicated to developing and applying scientific advances to health, agriculture, learning, and commercialization. Opened in 2008, HudsonAlpha’s vision is to leverage the synergy between discovery, education, medicine, and economic development in genomic sciences to improve the human condition around the globe. The HudsonAlpha biotechnology campus consists of 152 acres nestled within Cummings Research Park, the nation’s second largest research park. The state-of-the-art facilities co-locate nonprofit scientific researchers with entrepreneurs and educators. HudsonAlpha has become a national and international leader in genetics and genomics research and biotech education and fosters more than 40 diverse biotech companies on campus. To learn more about HudsonAlpha, visit hudsonalpha.org.


