The Friday poster session at the 2018 American College of Medical Genetics and Genomics (ACMG) was peppered with HudsonAlpha entries that shared new findings and new tools to enhance the analytical power of whole genome sequencing.
Donna Brown
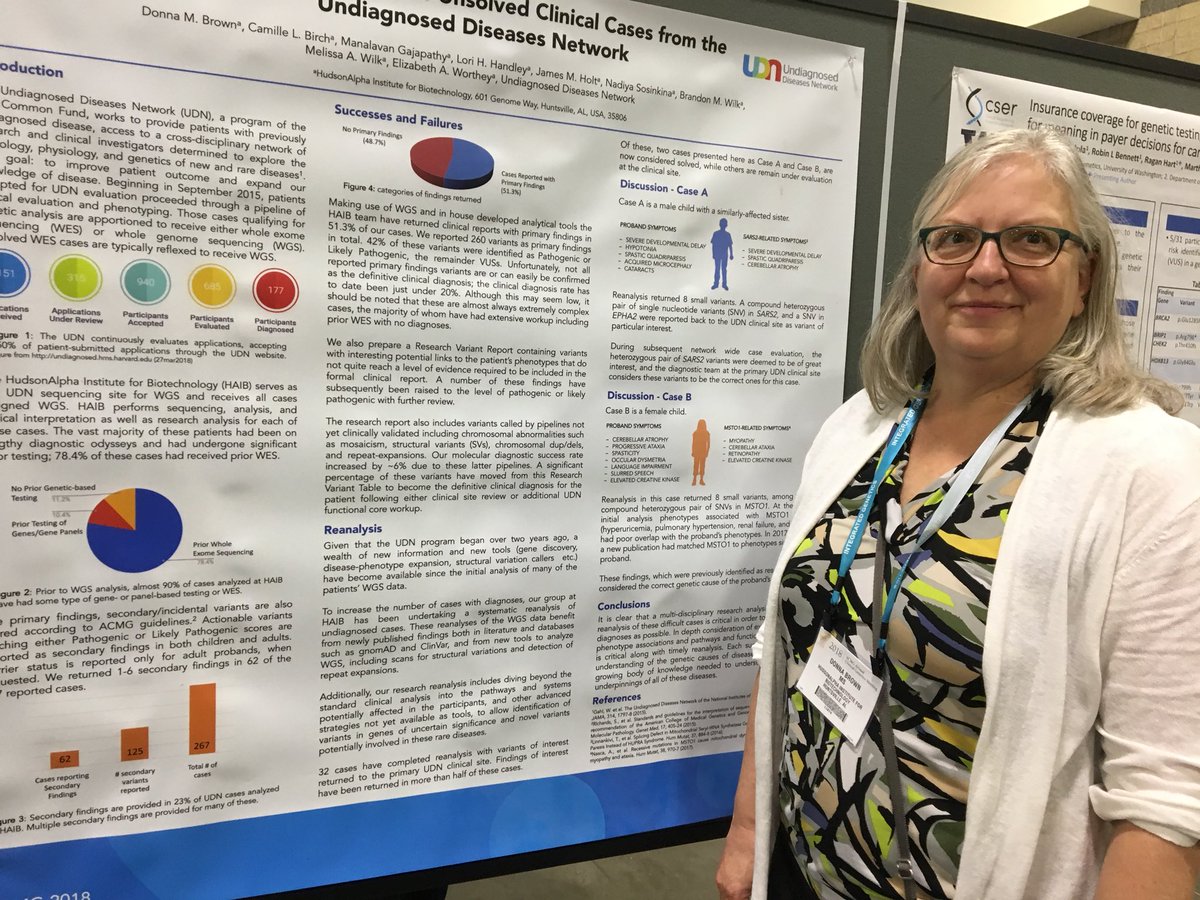
Donna Brown, clinical analyst in Software Development and Informatics, was awarded a blue ribbon for her poster exploring the power of reanalysis in cases sent for whole genome sequencing (WGS) from the Undiagnosed Disease Network (UDN). HudsonAlpha is the sequencing core for UDN. HudsonAlpha is beginning to reanalyze unsolved UDN cases that have already undergone WGS, and already has identified new variants in many cases. The group will continue the reanalysis project to get answers for patients.

Kelly East
Kelly East, genetic counselor, presented findings from the Information is Power initiative. The initiative offers free genetic cancer risk tests to people ages 28-32 in a five-county area of North Alabama. More than 2700 people have taken the test, and about 3% test positive for a genetic variant that increased breast or ovarian cancer risk. More than half those testing positive would not have qualified for cancer predisposition testing under the current standard of care. East’s poster discussed the challenge of providing ongoing care and genetic counseling for those individuals.
Mana Gajapathy, PhD, a postdoctoral researcher in Dr. Liz Worthey’s lab, presented a poster on his recent work integrating datasets into CODICEM, the analytical tool developed by the Software Development and Informatics group to analyze genomes. CODICEM already has integrated and frequently updates datasets including ClinVar, OMIM, and many others. The recent addition is UniProtKB, which is a collection of functional information on proteins (mainly amino acid sequence).
Matt Holt, PhD
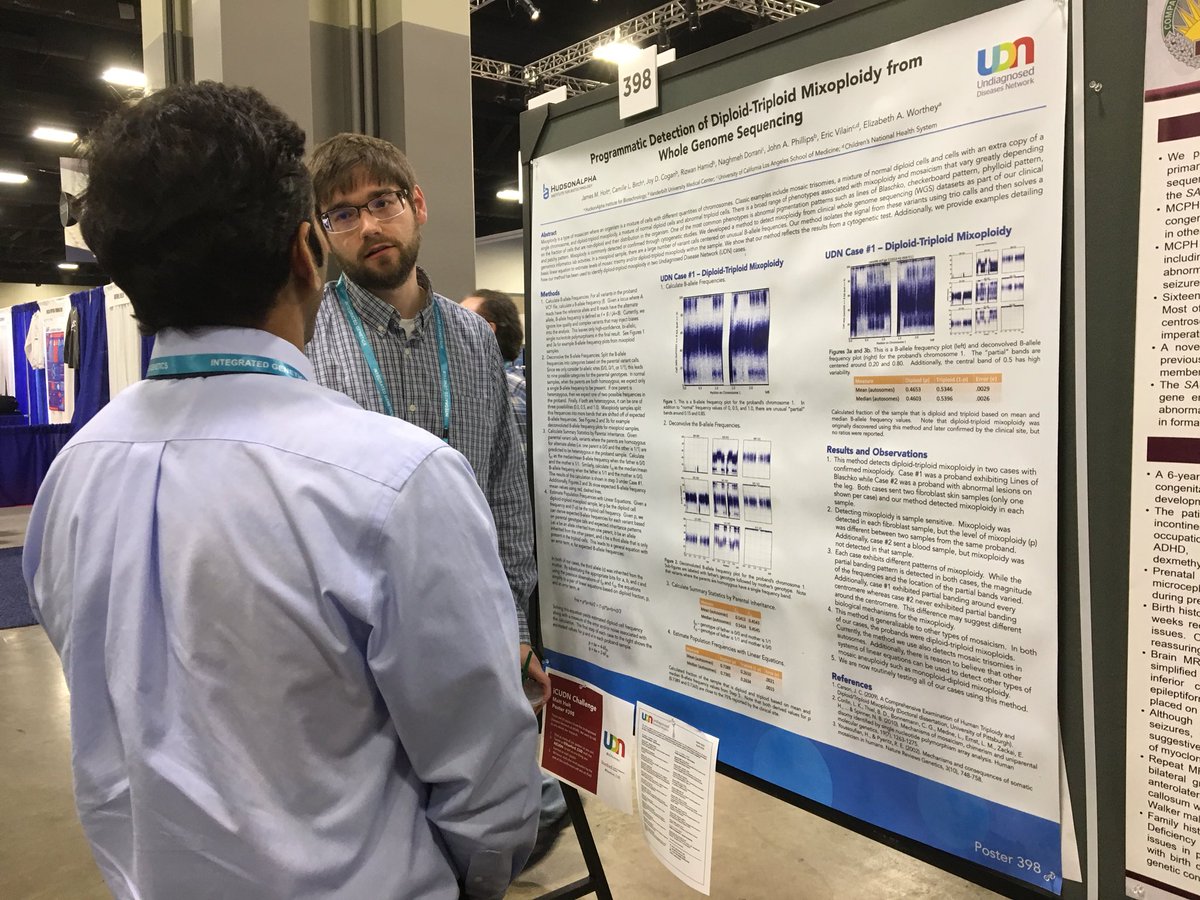
Matt Holt, PhD, postdoctoral researcher in Dr. Liz Worthey’s lab, shared his poster, “Programmatic Detection of Diploid Triploid Mixoploidy from WGS.” Diploid Triploid mosaicism is a disorder in which patients have some cells with two copies of each chromosome, but also cells with three copies of each chromosome. Cytogenic tests have typically identified diploid triploid mixoploidy, but Dr. Holt demonstrated a method by which whole genome sequencing can successfully identify (and quantify the percentage of triploid cells in) the disorder.
Adam Hott
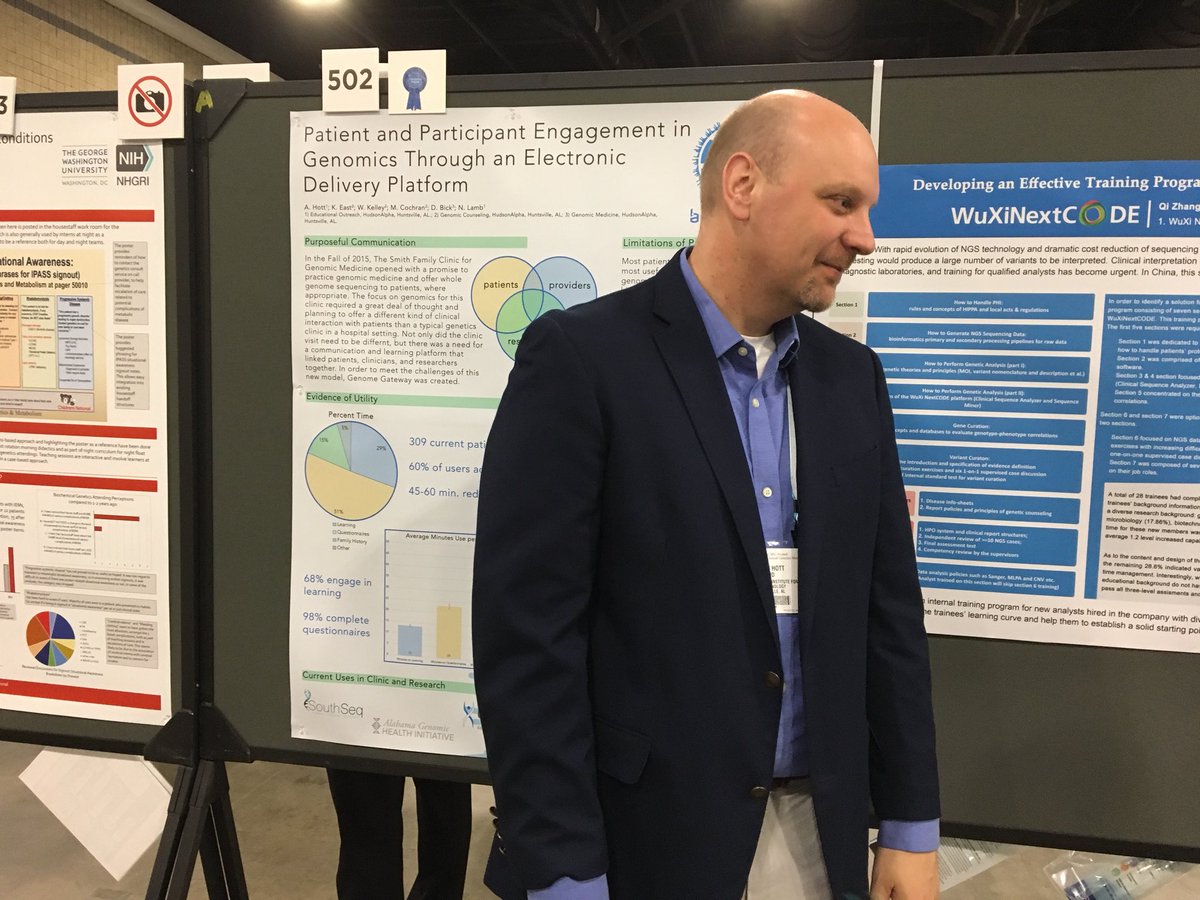
Adam Hott, EdD, a member of the HudsonAlpha Educational Outreach team and the digital applications lead, shared data on utilization of a unique patient tool at the Smith Family Clinic for Genomic Medicine called Genome Gateway. More than 300 patients have used Genome Gateway to input health history and other information and to utilize the learning tools. Dr. Hott found that more than 98% of patients utilize Genome Gateway to fill out health questionnaires – and that cut down the time spent in the clinical appointment by up to one hour for each patient.
Brandon Wilk, software developer in Dr. Liz Worthey’s lab, shared early data on a new tool being developed in the lab that matches phenotypes with the most likely gene candidates. The tool then prioritizes the list of likely genes for the analysts. The tool is designed to streamline the work of clinical analysts and find answers for patients more quickly.
Melissa Wilk, PhD, postdoctoral research in Dr. Liz Worthey’s lab, presented a case study to demonstrate the value of whole genome sequencing in cystic fibrosis (CF). The patient was diagnosed through newborn screening at birth and had an unusually severe case of the disease. Whole genome sequencing identified CFTR variants (associated with CF), both known and potential novel CF modifiers, and pharmacogenetic information predicted to affect CF therapies. The case demonstrates the value of WGS for researchers studying the disease.