An Everyday DNA blog article
Written by: Sarah Sharman, PhD, Science writer
Illustrated by: Cathleen Shaw
Viruses are tiny organisms that can wreak major havoc on human health worldwide, something that we have all witnessed first hand over the past year and a half. Some viruses, like influenza and HIV, are endemic foes, existing at constant levels in certain areas throughout the world. Others, like the SARS-CoV-2 virus, emerge anew and cause widespread outbreaks before they can be identified and controlled.
The ability to quickly identify and start combating viruses is paramount in minimizing death and devastation. Advances in genomic technologies have given scientists the ability to study viruses at a genetic level, allowing them to find ways to target the virus with drugs, make vaccines to prevent infection with the virus, and track and monitor the spread of viral outbreaks. Let’s learn more about how scientists use genomics to help combat viral pandemics.
What are viruses and why are their genomes so useful?
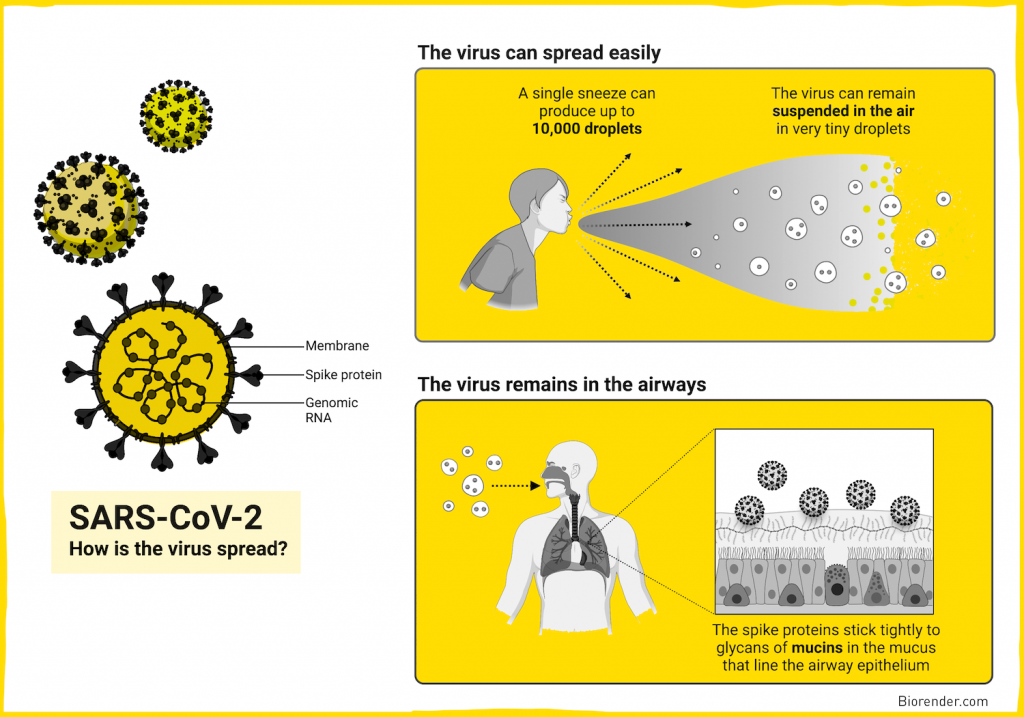
 Viruses are microbes that can infect nearly all forms of life. Viral particles consist of DNA or RNA wrapped in a protective layer of proteins. Viruses cannot replicate on their own and rely on host cells for replication. To successfully survive and reproduce, viruses must move through three stages: contact with a susceptible host, infection and replication within the host, and transmission to other individuals.
Viruses are microbes that can infect nearly all forms of life. Viral particles consist of DNA or RNA wrapped in a protective layer of proteins. Viruses cannot replicate on their own and rely on host cells for replication. To successfully survive and reproduce, viruses must move through three stages: contact with a susceptible host, infection and replication within the host, and transmission to other individuals.
When a virus enters a host (usually through the mouth, eyes, nose, or open wounds in the skin), it must quickly mount a successful infection before it is caught and killed by the host’s immune system. Many viruses have evolved specific interactions with host cells to achieve infection. For example, human flu viruses and SARS-CoV-2 viruses have proteins on their surface that bind to matching receptors on human respiratory cells.
Once inside the host cell, the virus hijacks the cell’s reproductive machinery and copies its own genetic material to make more viral particles. Replicating viral cells damage or weaken host cells which can cause symptoms of disease like fever, body ache, and other tissue specific symptoms. The new viral particles can be passed to another individual through bodily fluids, for example the flu is generally transmitted through sneezes or coughs.
Because viruses replicate so quickly, they can quickly develop random mutations in their DNA or RNA. Most mutations will have no effect, or will have a detrimental effect on the virus itself. However, a small proportion of the mutations may enable the virus a beneficial edge, allowing it to infect the host better, spread from individual to individual faster, or survive longer outside of a host for example.
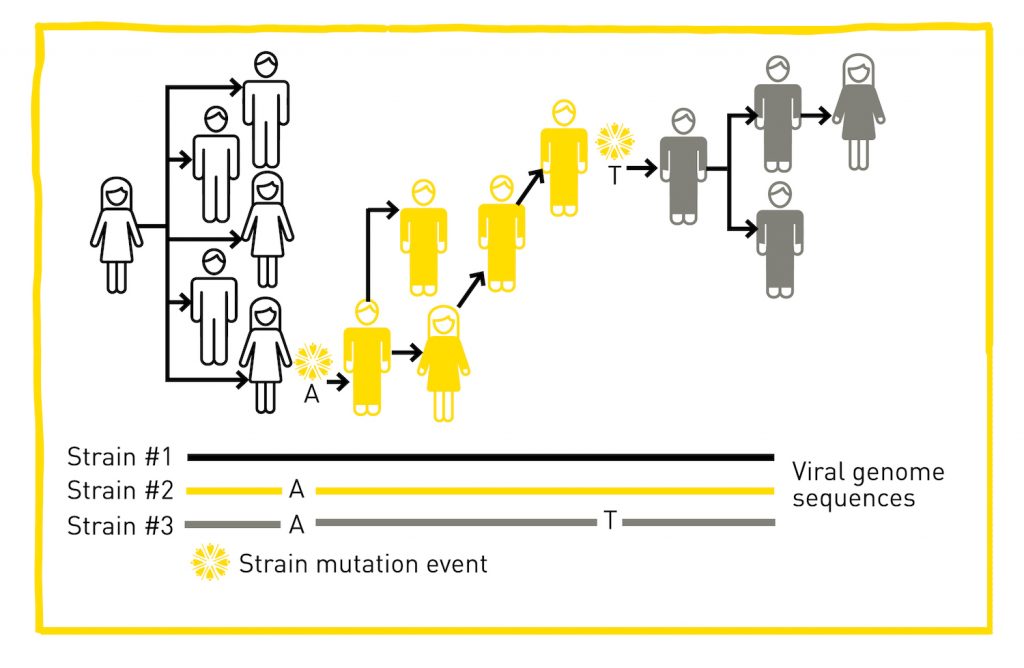 The influenza virus has proteins on its surface that are recognized by the immune system. Mutations in the genes that code for these proteins occur frequently which is the main reason that people can get the flu more than one time and why the flu vaccine composition must be updated each year. Scientists are able to determine how the influenza virus has changed thanks to genome sequencing.
The influenza virus has proteins on its surface that are recognized by the immune system. Mutations in the genes that code for these proteins occur frequently which is the main reason that people can get the flu more than one time and why the flu vaccine composition must be updated each year. Scientists are able to determine how the influenza virus has changed thanks to genome sequencing.
Genome sequencing is a process that helps scientists decipher the sequence of DNA and RNA nucleotides that make up each virus’s genetic code. By studying the genetic code, scientists can pinpoint mutations that viruses may have picked up over time. Knowledge gained from the genomic sequence of a virus can help scientists discover drugs and treatments to alleviate disease in infected individuals and produce vaccines to prevent infection with the virus.
Why is genomic surveillance important during an outbreak?
 In addition to helping treat and prevent viruses, scientists and epidemiologists also use viral genome sequencing for genomic surveillance to track and monitor the spread of viral outbreaks. By consistently sequencing viral genomes from many infected individuals, researchers can monitor how the virus changes over time, understand how these changes affect the characteristics of the virus, and use this information to better understand how it might impact an individual’s health.
In addition to helping treat and prevent viruses, scientists and epidemiologists also use viral genome sequencing for genomic surveillance to track and monitor the spread of viral outbreaks. By consistently sequencing viral genomes from many infected individuals, researchers can monitor how the virus changes over time, understand how these changes affect the characteristics of the virus, and use this information to better understand how it might impact an individual’s health.
Genomic surveillance is vital for detecting and characterizing new variants as they arise, a crucial part of the effort to control viral outbreaks. Detecting variants early in their lifecycle allows scientists to quickly determine important factors such as the rate the variant is spreading, the severity of disease, whether existing diagnostic tests will detect the variant, and whether existing vaccines will protect against the variant.
Successful genomic surveillance programs are fast and immediately make data publicly available to inform real-time decision-making by public health officials and vaccine manufacturers. The programs also consistently screen positive samples at multiple sites across a country or region to ensure the most comprehensive coverage of the viral landscape.
Tracking SARS-CoV-2 variants
Genomic surveillance has proven especially important during the ongoing COVID-19 pandemic. Countries with strong genomic surveillance programs in place, like the UK and Denmark, have been able to quickly detect emerging variants. For example, in the UK, the Alpha variant was detected early in its lifecycle. By studying the variant, scientists found several mutations in the spike protein that made it easier for the virus to infect human cells, making this particular variant more infectious.
Experts estimate that sequencing 10 percent of SARS-CoV-2 positive samples is highly effective at detecting the variants in a population, but randomly sequencing five percent of the positive samples may be sufficient. The United States is well under the targeted goal of sequencing viral genomes from five percent of the positive cases in the country.
As part of the effort to increase the number of positive samples that are sequenced in the United States, the HudsonAlpha Institute for Biotechnology’s Genome Sequencing Center is sequencing thousands of positive samples from across the state of Alabama. The project, led by Jane Grimwood, PhD, was initially funded through the CARES Act and is now funded through an award from Testing for America.
By sequencing viral genomes from SARS-CoV-2 positive samples throughout the state of Alabama, Grimwood and her team aim to help officials track variants of concern, track viral mutations over time, and detect any viral breakthrough that may occur after vaccination. The sequencing also helps to identify possible sources of new hotspots of infection in Alabama.
So far, the team has sequenced more than 1700 samples collected from 57 of the 67 counties in Alabama. Using the genome sequences, the scientists separated the samples into groups based on sequence variation and mutations. The scientists are also able to detect variants of concern, like the delta variant, among populations in the state to determine if there is an uptick in infected individuals.
Sharing this kind of data with state and national databases helps experts better understand the genomics and patterns of SARS-CoV-2 transmission across the United States by adding an Alabama perspective to the picture.