Genome Editing and CRISPR
Tools that directly modify genetic sequences allow scientists to explore the functional impact of DNA mutations as well as engineer changes that create drug-producing bacteria, disease-resistant crops or life-saving genetic therapies. These approaches are analogous to the “find and replace” feature in word processing software – they scan the genome for a specific sequence and then make a targeted modification within that sequence.
All genome editing tools rely on some sort of programmable nuclease – an enzyme guided to a DNA sequence of interest in order to cut across the DNA strand. This cut triggers a DNA repair process that can knockout (disrupt) the genetic instructions or replace them with a different set of infor-mation. There are three programmable nucleases – zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeat Cas-9 system (CRISPR-Cas9). The CRISPR-Cas9 system has proven especially powerful and has quickly become a mainstay of genome editing protocols in microbes, plants and animals.
Genome editing tools have provided researchers with an unprecedented ability to modify cells. This leads to applications in basic research, agriculture and human disease therapy. However, programmable nucleases are not without their challenges. They can be difficult to correctly deliver to the appropriate cells, the frequency of DNA modification can be very low, and they occasionally cut DNA at “off target” sites. Techniques that increase specificity and efficiency have been developed to address these challenges, but additional tweaking is still needed.
Genome editing has shown immense promise – the first wave of modified crops and human therapies are moving from the laboratory to the field and clinic. A number of regulatory and safety hurdles must still be cleared, and there are many ethical, social and policy implications waiting to be addressed.
The CRISPR-Cas9 method has many applications in basic research, agriculture, drug development and eventually treating humans with genetic diseases.
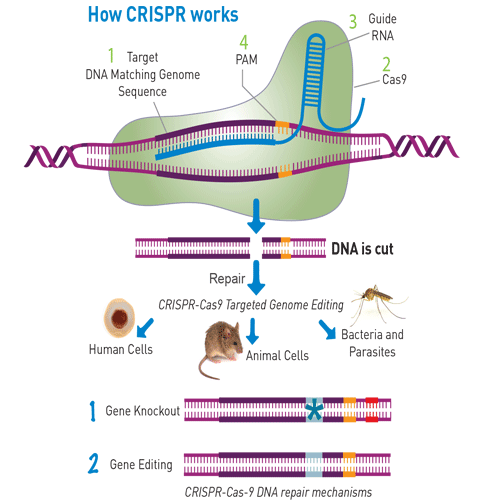
There are four key components of a CRISPR-Cas9 system:
1. Target DNA: This is the region of the genome to be modified.
2. Cas9: This bacterial enzyme unzips and cuts the target DNA. To date, most approaches use the Cas9 protein found in the bacteria Streptococcus pyogenes.
3. PAM sequence: PAM stands for Protospacer Adjacent Motif. It is part of the target sequence DNA and is one of the factors that is required to define the cutting site.
4. Guide RNA: A short fragment of RNA binds to Cas9 and contains a recognition sequence that matches the target. With different guide RNA sequences, the Cas9 enzyme can be directed to recognize almost any DNA sequence. The guide RNA leads Cas9 to the desired location in the genome, binds the target sequence and triggers Cas9 to cut both strands of target DNA. This double stranded break provides the opportunity to edit the genome.