Part 1: The birth of DNA sequencing
by: Sarah Sharman, PhD, Science writer
The first National DNA Day was celebrated on April 25, 2003 to commemorate both the successful completion of the Human Genome Project and the discovery of DNA’s double helix in 1953. The goal of DNA Day is to offer students, teachers and the public an opportunity to learn about and celebrate the latest advances in genomic research and explore how those advancements might impact their lives.
So, what are we waiting for? Let’s celebrate DNA Day by diving into the history of DNA sequencing technology and learning how advances in technology impact our lives today.
The birth of DNA sequencing
Inside each plant or animal cell is a code of instructions for life, called a genome. Genome is just a fancy word for the collection of all of the deoxyribonucleic acid, or DNA, in an organism. DNA is made up of four chemical bases represented by the letters A, C, G and T, that combine in different three base combinations (such as ATG or TGG) to form amino acids which are the building blocks of proteins. There are more than 20,000 proteins that work together to perform all of a human’s functions.
Being able to detect and analyze changes in an organism’s DNA is important for answering questions about health, disease, and other biological phenomena. However, we have not always been able to easily identify the precise order of DNA bases, called a DNA sequence. In fact, rapid sequencing technology did not come about until almost twenty years after James Watson and Francis Crick, along with Rosalind Franklin and Maurice Wilkins, proposed that DNA exists as a three-dimensional, double-helix structure.
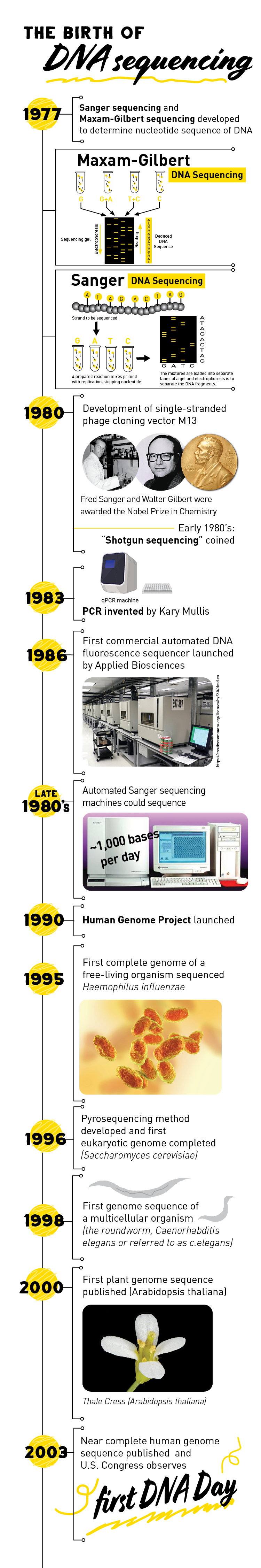
Around 1977, two groups independently developed rapid methods to determine the sequence of DNA, decoding hundreds of bases in a day. The methods, called Maxam-Gilbert sequencing (chemical sequencing) and Sanger chain termination sequencing, represent the first generation of DNA sequencing technology.
The Maxam-Gilbert method uses chemical processes to break the bonds between DNA bases, creating many shorter fragments of DNA. Each different chemical process breaks the bond between specific DNA bases. By running each chemical process in a separate tube, the scientists know which base was at the end of the terminated fragment. The fragments are then sorted by size to determine the order of the DNA bases.
Sanger sequencing, like many other sequencing technologies to date, is based on the natural process of DNA replication. During DNA replication, the DNA double-helix is unzipped into two single strands. Then new nucleotides are added one by one to each strand by an enzyme called DNA polymerase. As polymerase is adding new nucleotides, the strands are zipped back up, creating two complete DNA helices.
In Sanger sequencing, special bases are randomly incorporated by DNA polymerase into the strand it is copying. The special bases stop DNA polymerase from adding any more bases, resulting in different sized pieces of DNA. Similar to Maxam-Gilbert sequencing, the fragments are sorted by size and the sequence of the bases is determined based on the presence of a label attached to the special bases.
Both Maxam-Gilbert and Sanger sequencing were immediately adopted by scientists. Maxam-Gilbert was the most popular sequencing method at the time. However, due to the complexity of the method and the harsh chemicals required for the reaction, Sanger sequencing ultimately surpassed Maxam-Gilbert sequencing in popularity.
Improving Sanger sequencing: shotgun sequencing, automated machines and more
While Sanger sequencing quickly became the gold standard of DNA sequencing, it could only be used for short DNA strands of 100 to 1000 base pairs (for reference, the entire human genome is made up of 3.1 billion base pairs). In 1979, a new technique called ‘shotgun sequencing’ was introduced to solve this problem.
During shotgun sequencing, DNA is broken up into many smaller segments, which are then inserted in bacterial vectors to make many copies of the segment. The segments are then sequenced using Sanger sequencing to obtain multiple sequences, called reads. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence.
Other technologies were developed that helped improve the accuracy and speed of Sanger sequencing and shotgun sequencing. For example, the development of the single-stranded M13 phage cloning vector in 1980 and polymerase chain reaction in 1983 allowed scientists to amplify, or copy, DNA by several orders of magnitude.
Two improvements that facilitated the automation of Sanger sequencing were the introduction of dye-labeled terminators, which allowed for one rather than four sequencing reactions, and the development of capillary electrophoresis, which replaced gel-based electrophoresis and simplified the steps of extraction and interpretation of the fluorescent signal.
The first commercial DNA sequencer, which automated many of the Sanger sequencing steps, was launched by Applied Biosciences in 1986. Scientists could now simply feed prepared DNA into a machine and view the results of fluorescence-based reactions on a screen. By the late 1980’s, automated Sanger sequencing machines could sequence about 1,000 bases per day.
Sequencing the tree of life
Over the course of the next decade, improvements to Sanger sequencing and shotgun sequencing technologies would allow scientists to sequence the genomes of many different, increasingly complex organisms. The first complete genome of a free-living organism, the bacterium Haemophilus influenzae, was completed in 1995, followed by the first eukaryotic genome (the yeast Saccharomyces cerevisiae) in 1996, the first genome sequence of a multicellular organism (the roundworm Caenorhabditis elegans) in 1998, and the first plant genome (Arabidopsis thaliana) in 2000.
Perhaps the most monumental sequencing accomplishment of the 21st century was the completion of the first near complete human genome sequence. It was sequenced through an international, collaborative effort called the Human Genome Project. The Human Genome Project was a publicly funded project initiated in 1990 with the objective of determining the DNA sequence of the entire human genome within 15 years.
The international team, which included more than 2,800 researchers at 20 institutions across the United States, the United Kingdom, France, Germany, Japan and China, delivered a near-complete human genome two years ahead of schedule in April 2003. The genome showed that humans have about 3.1 billion nucleotides that make up about 20,000-25,000 human protein-coding genes. Research Highlight #1 describes how HudsonAlpha scientists contributed to the Human Genome Project.
To learn about the next stage of genome sequencing technology, stay tuned for Part 2 of this series.

The Human Genome Project
Because the human genome is so large, the international project split the sequencing workload up between many different institutions, dividing it based upon chromosomes. At the time, human chromosomes were too large to sequence whole, so scientists had to break them up into more manageable sizes using a hierarchical shotgun strategy to sequence them.
Each chromosome was fragmented into shorter sequences of about 150,000 bases which were inserted into bacterial artificial chromosomes (BACs) to generate vast numbers of each short sequence. DNA purified from the BACs was used as a template for automated Sanger sequencing. The short sequence reads were overlapped with their neighbors and assembled into larger, contiguous stretches of DNA known as contigs. Ideally, each chromosome would be represented by a single contig, but the first draft consisted of 1,246 contigs.
HudsonAlpha President and Science Director Richard M. Myers, PhD, along with Faculty Investigators Jane Grimwood, PhD, and Jeremy Schmutz, were part of the Stanford Human Genome Center team that were involved in sequencing the 320 million base pairs of human chromosomes 5, 16 and 19. More specifically, they performed finishing and quality analysis on the chromosomes. This means that they manually reviewed the sequences one base pair at a time, running computational analysis and ordering new sequences to fill gaps, reconcile sequence repeats, and make the sequence cleaner overall.